Integrating mutation, copy number, and gene expression data to identify driver genes of recurrent chromosome-arm losses

A data-driven method to optimize the clinical scales for Parkinson’s disease, improving its ability to track disease progression.

A novel two-stage clinical trial design that meets regulatory requirements while enhancing real-world outcomes and fairness.

4-class classifier of metagenome contigs using machine learning and assembly graphs

Integrating gene expression and DNA methylation datasets for disjoint sample sets.

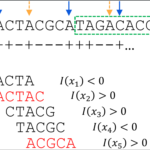
Selecting minimizers based on minimum decycling set.
CT-FOCS - A database of cell type-specific enhancer-promoter links

Long-read mapping using syncmers.
Accurate module discovery using gene activities and a network
Dataset adaptive minimizer order generator for binning applications

3-class classification of metagenome assemblies

Multi Omic clustering by Non-Exhaustive Types

Complete plasmid assembly in metagenomes

Empirically correcting the functional significance of GO terms in modules

PlasClass - Plasmid sequence classifier

EXPANDER - A one stop-shop platform for all analysis steps of gene expression data.

PROMO - a tool for analysis and visualization of large multi-omic and clinical datasets.
NEMO: Neighborhood based multi-omics clustering algorithm

Adeptus - A database and tools for multi-disease analysis.

Faucet: streaming de novo assembly graph construction

FOCS: A database of human and mouse enhancer-promoter links

Recycler - detecting plasmids from de novo assembly graphs

DOCKS - A software for designing compact universal k-mers hitting set.

RichMind - Analysis of enrichment within neuroimaging findings

Motif finding from high-throughput SELEX

ElemeNT - Tool for detection of core promoter elements.

BARCODE - A fast lossless read compression tool based on Bloom filters.

RAP - A software for inferring binding site motifs from protein binding microarrays.

HiDe - A software for inferring highways of horizontal gene transfer.

ShortCAKE - Designing a shortest DNA sequence that covers all k-mers, exploiting reverse complementarity.

AMADEUS - Platform for de novo motif discovery in DNA sequences.

Morph - A webtool and software for ranking candidate genes in pathways.

ALLEGRO - Software for simultaneous discovery of cis-regulatory motifs and their associated expression profiles.

ModEnt - A tool that reconstructs gene regulatory networks from high throughput experimental data.

STACK - Analyzing statistical associations among cancer karyotypes.

FAME - A tool for Functional Assignment to MicroRNAs via Enrichment

TORQUE - Topology-free querying of protein interaction networks.

MetaReg - A tool for modeling of biological systems

HYDEN - A software for designing degenerate primers.

SPIKE - (previously SHARP). Signaling pathways integrated knowledge engine.

GEVALT - An integrated software for genotype analysis.

PSAT - Evaluating the significance of association scores in whole-genome cohorts with stratification.

Cofactors - Linking transcription factors and chromatin modifiers.
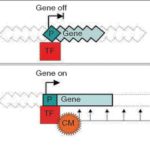
MATISSE - A tool for joint analysis of interaction networks and gene expression data.

RAT - A software for rapid association tests.

Cisprof - Evolutionary mechanisms underlying conservation and evolvability in regulatory networks.

SIMBA - System level analysis of yeast by exploring of large-scale biological compendia.

SAMBA - Module discovery in highly diverse genome-wide datasets using biclustering.

GREAL - software for the graph realization Problem.
PREVO - An analysis of the network of selection forces in the evolution of yeast cis-regulation

PRIMA - A program for finding transcription factors whose binding sites are enriched in a given set of promoters.
PIVOT - Java-based protein interactions visualization tool.

