|
MetaReg is a Java application aimed to provide the biologist with an intuitive graphical
interface for both definition and analysis of system models. The application makes it
possible to navigate the model network and compare its predicted and observed behavior.
MetaReg features:
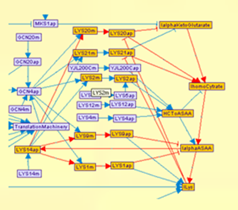
- Model definition: A MetaReg model consists of a set of biological variables and their
discrete regulatory logics. The biologist can easily organize the model in a coherent and hierarchical
structure by using the fully interactive model canvas. Variables can be added to the model and
automatically linked to known databases (SGD, NCBI Gene). The logic definition is facilitated by
either scripting or hierarchical construction capabilities.
|
|
-
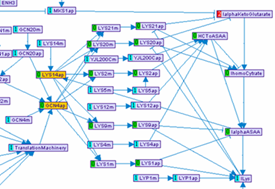
Performing simulations on the model: In order to test the compatibility of the model with the
expected behavior of the network in response to different stimulations, simulations can be performed.
Given a set of stimulations and perturbations, the logical states of all model variables are inferred
as described in [1]. In case several modes are feasible, the user can navigate among them, visually
analyzing the possible state of the system.
|
|
- Model predictions: After the model is specified and relevant measured data are loaded, the user
can execute
the prediction algorithms described in [2]. The user can control the full parameter setting for each of
the supported methods (Gibbs Sampling, Mode Instantiation, Loopy Belief Propagation and Exact Inference).
|
|
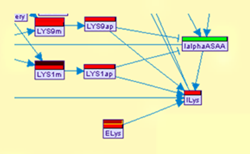
- Observed and predicted data comparison: For each experimental condition, the application predicts
the state of each variable, which can then be compared to its measured value. One can view either the measured
states, or the predicted states, or the discrepancies between them. This can be done through (1) display of the
states on the model canvas or (2) a matrix view display. In the matrix display of discrepancies, each
cell contains a color representation of both states, along with a color-coded representation of the
discrepancy be-tween the states. This view allows a simple detection of discrepancy
“hot-spots” in which the model fails to explain the data.
|

|
- Model refinement: Once the parents of a variable in the model is set, its logic can be refined using
a probabilistic refinement engine. The refined logic is augmented with significance flag for every possible
combination of the variable's parents.
|
|
References
-
MetaReg: A platform for modeling, analysis and visualization of biological systems using large-scale experimental data..
I. Ulitsky, I. Gat-Viks, R. Shamir
Genome Biology (2008) Vol. 9 No. R1 
-
Factor graph network models for biological systems.
I. Gat-Viks, A. Tanay, D. Raijman, R. Shamir
Proc. RECOMB 2005 , pp. 31-47, Lecture Notes in Bioinformatics 3500, Springer, Berlin, 2005.

-
Modeling and Analysis of Heterogeneous Regulation in Biological Networks
I. Gat-Viks, A. Tanay and R. Shamir
Journal of Computational Biology.
Vol. 11 No. 6 pp 1034--1049 (2004).
Downloading MetaReg
A version of MetaReg is freely available for academic use, and can be downloaded here.
The probabilistic inference and refinement engines are currently fully operational on the Windows environment. On Linux environment, MetaReg is operational, but still in beta status .
Users interested in commercial applications should contact Ron Shamir
In order to use MetaReg the Java Runtime Environment (JRE) must be
installed on your computer. MetaReg is designed to work with JRE
v1.5 or later, but it is strongly recommended that you download and install the latest version of the JRE available
for your operating system. Get the newest version of the JRE at: http://www.java.com/
MetaReg online manual
A manual for the use of MetaReg is available here. If you find any features
not comprehensively explained please contact us.
Example model
- A simple small model can be found here here.
- The leucine biosynthesis MetaReg model can be found here.
Both models already contain some experimental data and both are available in the examples/ directory in the application root.
Software updates
The following changes have been recently introduced to the software:
| Date | Change |
| 16/01/2008 | Bug fixes and extended species support |
The people behind MetaReg
MetaReg is developed in Prof. Ron Shamir's
Computational Genomics Laboratory at the School of Computer Science,
Tel Aviv University, Israel.
The developers are Igor Ulitsky,
Irit Gat-Viks and
Ron Shamir.
Feedback
For any questions, bugs and suggestions, please contact: Igor Ulitsky.
Support
This research was supported by the EMI-CD project that is funded by the European Commission
within its FP6 Programme, under the thematic area “Life sciences, genomics and biotechnology for
health”, contract number LSHG-CT-2003-503269.
MetaReg Home
|







