MATISSE - identifying modules using gene expression and interaction networks
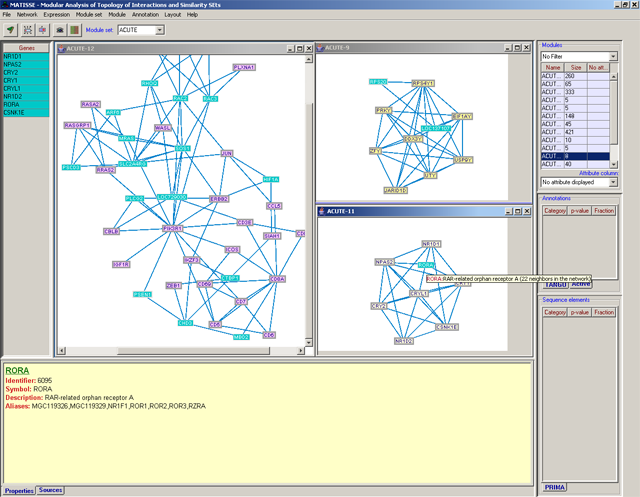
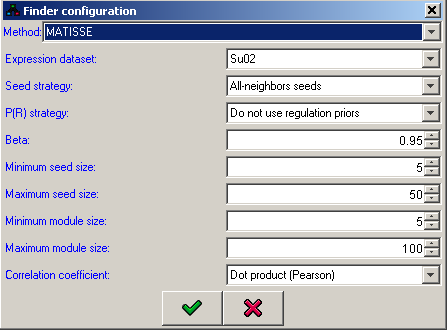
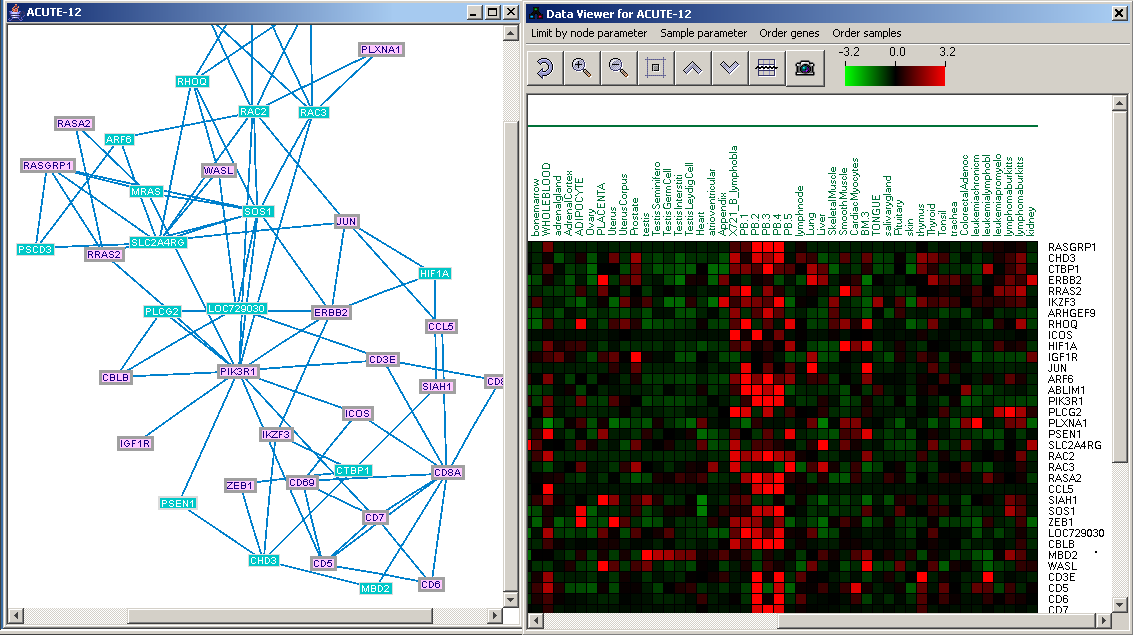
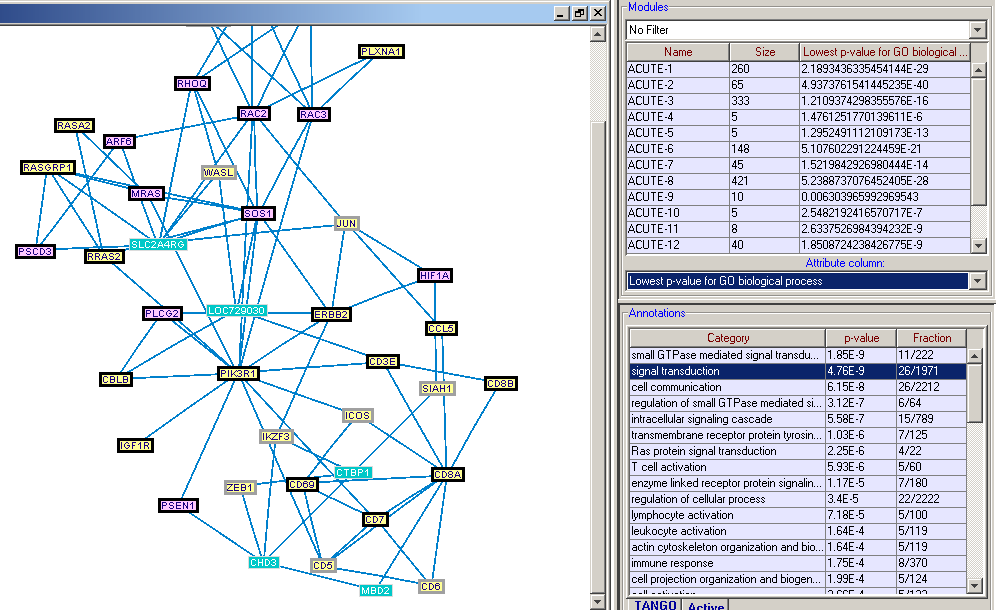
MATISSE (Module Analysis via Topology of Interactions and Similarity SEts) is a program for detection of functional modules using interaction networks and expression data. A functioncal module is a group of cellular components and their interactions that can be attributed a specific biological function.MATISSE implements several algorithmic engines that analyze together expression and network data. Each of the algorithms addresses a different research question and scenario. The software is activated using a friendly graphical user interface.
MATISSE currently supports the analysis of data from the species H. sapiens, M. musculus, S. cerevisiae, D. melanogaster, and C. elegans.

 Download MATISSE v1.1 for Windows and Linux (beta)
Download MATISSE v1.1 for Windows and Linux (beta)