| Home |
| Algorithms |
| Screenshots |
| User manual |
| Hands on |
| Download |
| Citations |
| References |
| version history |
| Acknowledgements |
| Google Forum |
| Contact Us |
| Home | |
|
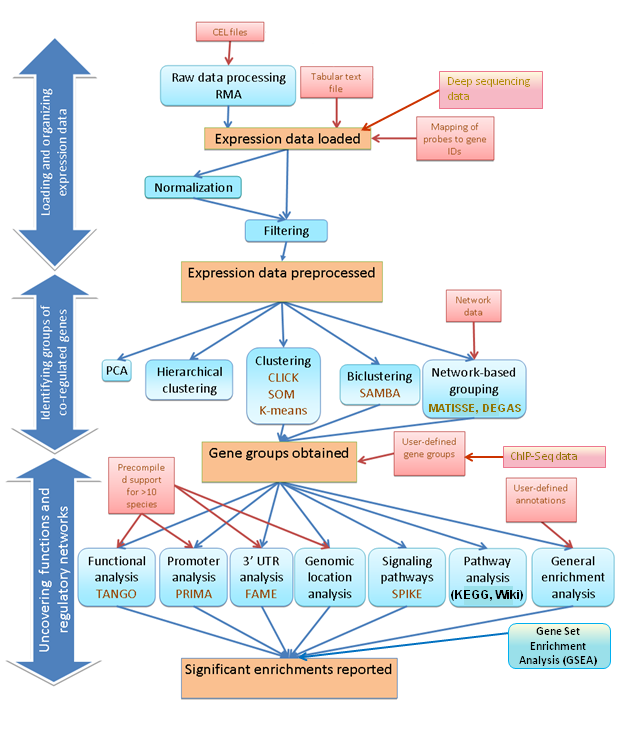
Overview:
EXPANDER provides support for
analysis of human, mouse, rat, chicken,
fly, zebrafish, C. elegans, S. cerevisiae, S. pombe,
arabidopsis, tomato, rice, listeria, A. fumigatus and E. coli. |
News and Notes
-
December 2019: "Basic tools for computational genomics - first steps with EXPANDER"
A tutorial by Tom Hait at The Weizmann Institute, on Sunday 29 December 2019, 9:30-14:30, Feinberg building, room B. Registration is required. June 2019: New Expander paper published
Hait et al. Journal of Molecular Biology https://doi.org/10.1016/j.jmb.2019.05.013.-
March 2019: Expander in Russian
The Expander website has been translated to Russian. See here -
October 2017: Expander 7.2 version is available for download
Version includes updated database for most organisms including human, mouse, and rat, a support for JASPAR PWMs in Prima and Amadeus, and an improved visualization with explanations on the computed p-values.Check it out! -
May 2016: "Basic tools for computational genomics - first steps with EXPANDER"
A tutorial by Tom Hait at TAU Bioinformatics Workshops Series 2015-16 , on Monday 23 May 2016, 10:00-14:00, Sherman building, room 09. Registration is free but required: click here -
April 2016: Expander 7.11 sub-version is available for download
Version includes "ChIP-Seq Analysis" menu item for performing all ChIP-Seq related analyses in Expander, option to fetch ChIP-Seq peaks sequences and performing on them AMADEUS motif finding, improved Expander visualization and an additional ranking option in GSEA.Check it out! -
January 2016: Expander 7.1 version is available for download
Version includes full AMADEUS visualization for motif analysis, DESeq2 option for differential expression analysis of RNA-Seq count data and minor changes in GSEA . Check it out! -
October 2015: Expander 7.01 sub-version is available for download
Version includes an improved visualization and a bug fix in GSEA, improved preproccesing condition re-ordering and an improved RankSum differential expression test . Check it out! -
October 2015: Expander workshop in Ghent, Belgium
A workshop on Expander use will be held on October 29 2015 at the Department of Plant Systems Biology, VIB, Ghent, Belgium. The workshop will be conducted by Dr. Oren Tzfadia. -
May 2015: Expander version 7 is available for download
Version includes support for ChIP-Seq data analysis, support for simultaneous analysis of more than one data set, improved network visualization using Cytoscape. Check it out! -
August 2014: Expander 6.5 sub-version is available for download
Version includes better support for NGS data analysis, improved heat-map visualization, post-clustering cleaning tool and an improved version of the Tango functional analysis procedure. Check it out! -
April 2014: "Advanced Computational Genomics Tools: Network Analysis and Gene Regulation "
A workshop presenting various network analysis and gene regulation tools will be held at TAU Bioinformatics Workshops Series 2013-14 , on Tuesday April 8th 2014, 10:15-13:00, Sherman building, room 09. Registration is free but required and open till 17.3.14: click here -
March 2014: "Basic tools for computational genomics - first steps with EXPANDER"
A tutorial by Adi Maron-Katz at TAU Bioinformatics Workshops Series 2013-14 , on Tuesday 18 March 2014, 10:15-13:00, Sherman building, room 09. Registration is free but required and open till 17.3.14: click here -
January 2014: Expander 6.3 sub-version is available for download
Version includes several improvements in GSEA analysis and ability to save all analysis results to txt files and figures at once -
November 2013: Expander 6.2 sub-version is available for download
Among the new features in version 6.2: Gene sets enrichment analysis (GSEA) and integration of AMADEUS software for motif discovery -
June 2013: Expander 6.1 sub-version is available for download along with up-to-date organisms-specific data files
Among the new features in Version 6.1: Wiki-pathways based enrichment analysis, automatic installation of required R scripts and an option to load multiple labels for experimental condition annotations. -
May 19th 2013: Expander hands on workshop
Advanced tools from the Shamir lab for analyzing microarrays, cis-regulation, and signaling pathways . TAU Bioinformatics Unit. - Some of our tools are now availble for download in batch mode (see download page)
-
January 2013: Expander 6.06 sub-version is available for download along with up-to-date organisms-specific data files
Among the new features in Version 6.06: Updated organism data for all supported species , newly added cdf files to facilitate cel file preprocessing and an improved coloring scheme for enrichment histograms -
April 22nd 2012: Expander hands on workshop
Advanced tools from the Shamir lab for analyzing microarrays, cis-regulation, and signaling pathways . TAU Bioinformatics Unit. -
December 2011: Expander 6.0 version is available for download
Among the new features in Version 6: Degas algorithm for network based clustering, ISA algorithm for bi-clustering , Hierarchical clustering tree partition , and support for similarity data. Check it out! -
July 4th 2011: Expander hands on workshop
Advanced tools from the Shamir lab for analyzing microarrays, cis-regulation, and signaling pathways . Bar-Ilan Bioinformatics Unit. -
April 12 2011: Expander hands on workshop
Advanced tools from the Shamir lab for analyzing microarrays, cis-regulation, and signaling pathways . TAU Bioinformatics Unit. -
March 31st 2011: Support for analysis of rice data added with release of subversion 5.2.3
-
May 2010: Programmer needed
We seek a Java pro to join the Expander development team. details - January 2010: Expander protocol published:
Ulitsky et al. Nature Protocols Vol 5, pp 303 - 322, 2010 .
Highlights of Expander 7:
- Updated database for most organisms
- Full AMADEUS visualization
- DESeq2 tool for performing differential expression analysis on RNA-Seq data
- ChIP-Seq data analysis support
- Support for simultaneous analysis of more than one data set (same organism)
- Addition of Cytoscape interface for improved network visualization
- Improved GSEA visualization
* Click here to view versions history.
The people behind EXPANDER:
Expander is developed in Prof. Ron Shamir's Computational Genomics Laboratory at the School of Computer Science, and in Dr. Rani Elkon's Mechanisms of Gene Regulation group at the Faculty of Medicine, Tel Aviv University, Israel. It is part of an ongoing effort to develop analytically sound and practical tools for complex analysis of gene expression and other high throughput genomic data. The following people have taken part in developing EXPANDER:
- Adi Maron-Katz
- Tom Hait
- Gabrielle Ecanow
- Aviv Steiner
- Seagull Shavit
- Dr. Chaim Linhart
- Dr. Igor Ulitsky
- Dr. Amos Tanay
- Dr. Roded Sharan
- Michael Gutkin
- Israel Steinfeld
- Naama Arbili
- Deepak Ajwani
- Erez Hartuv
- Dorit Sagir
- Akshay Krishnamurthy
- Prof. Richard M. Karp
- Prof. Yosef Shiloh
- Dr. Rani Elkon
- Prof. Ron Shamir