The goal here is to detect groups of genes that demonstrate similar expression patterns and are also highly connected in a given interactions network.
Before operating this tool, an interactions network in .SIF format must be loaded. This can be done by selecting Data>>Load network.
Network based grouping is performed by Expander using the Matisse algorithm (for details see the References section). The groups detected by Matisse are referred to as “modules” and may contain also genes that exist in the network, but are not present in the filtered GE data (referred to as “Back nodes”).
To use the more advanced, stand-alone version of MATISSE (with higher flexibility), please refer to the Matisse home page.
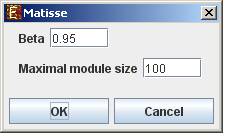
In order to apply the Matisse algorithm to the data select Grouping>>Network >>Matisse. The following dialog box will appear:

It enables the configuration of some of the parameters for the algorithm:
|
Field |
Description |
|
Beta |
The fraction of gene pairs that are expected to be strongly co-expressed in each module |
|
Maximal module size |
The maximum size for a detected module. |
Upon clicking ‘OK’ in the dialog box, the Matisse algorithm is operated on the dataset.
After running the algorithm, the solution is displayed in a new tab, which is added to the main window. The view is similar to the clustering results display. In the display, back nodes (genes that appear In the network, but not in the GE data) are marked in yellow.
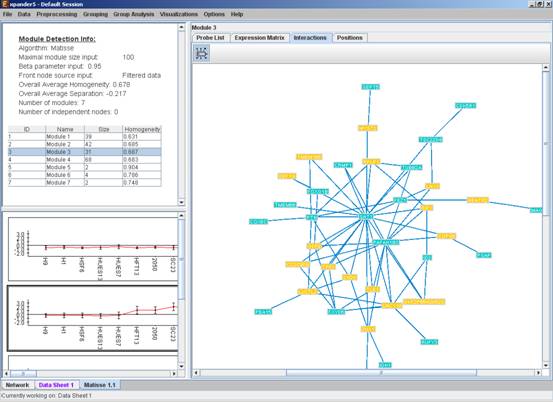
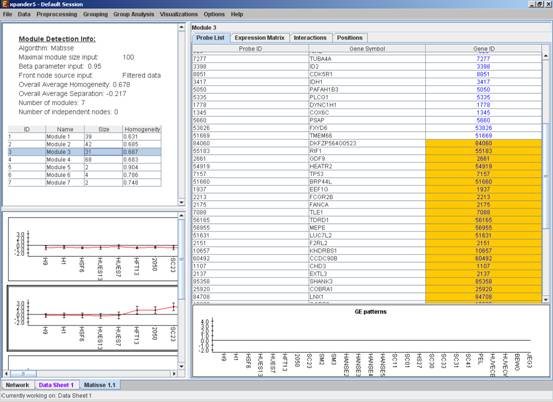
After performing group analysis (for details see the Group Analysis Tools section), if enrichment has been detected in the selected module, the corresponding histogram and analysis information are added to the single module view, and a column is added to the expression matrix display for each enrichment class, stating for each probe, whether it belongs to that class.
A network-based grouping solution can be saved using the Grouping >> Network >> Save Solution, and reloaded using the Grouping >> Network >> Load Solution. For a format of the solution file, please refer to the File Formats section: