
ModMap - Detecting maps of modules in biological networks
ModMap is a tool for advanced analysis of biological networks. Given two biological networks, Modmap seeks a set of modules, each representing strongly connected nodes in the first network, that are also strongly interconnected in the second network. The output is a map of the modules and their links. Unlike clustering, where the clusters are built based on one network, here the two networks are used simultaneously to build the map.ModMap can also be used for differential correlation analysis. It is based on ideas used for that goal in DICER, but it is more accurate and more robust.
ModMap was developed by David Amar in Ron Shamir's Computational Genomics group at Tel Aviv University.
Module maps: example
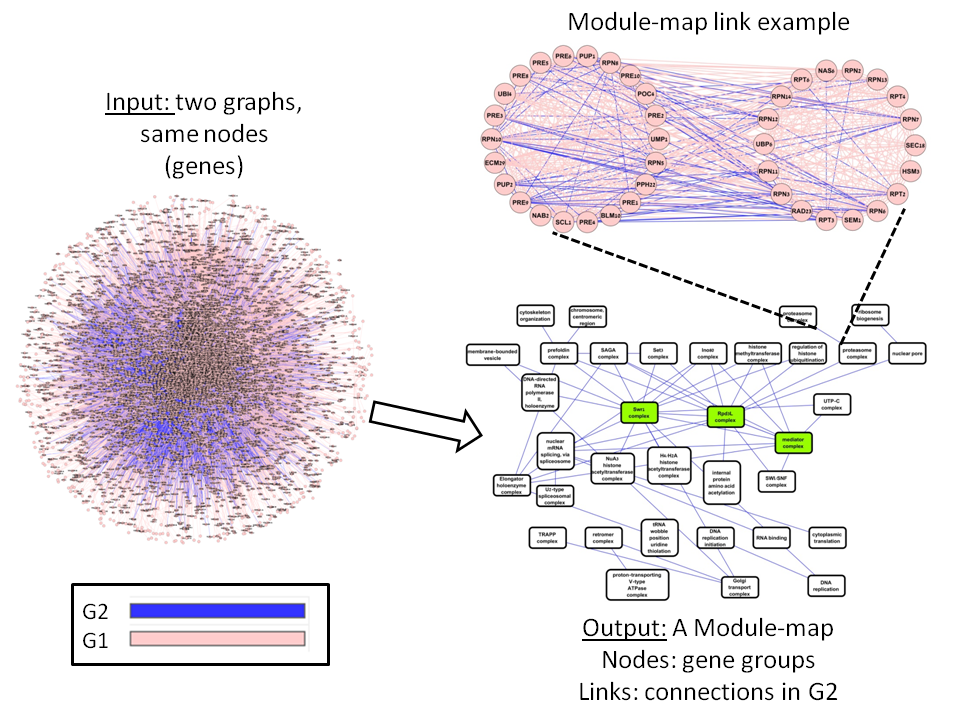 |
|
Get the software
ModMap is a research tool, still in the development stage.Hence, it is not presented as error-free, accurate, complete, useful, suitable for any specific application or free from any infringement of any rights. The Software is licensed AS IS, entirely at the user's own risk.
Below we explain how to run the executables. The source code is available here
Analyzing a pair of unweighted graphs
Input networks
The input files with the graphs for the unweighted tool are tab delimited with a single line for each interaction, i.e., geneA geneBExample of input files can be found here
These files represent two graphs in which the following map was planted (with other nodes):
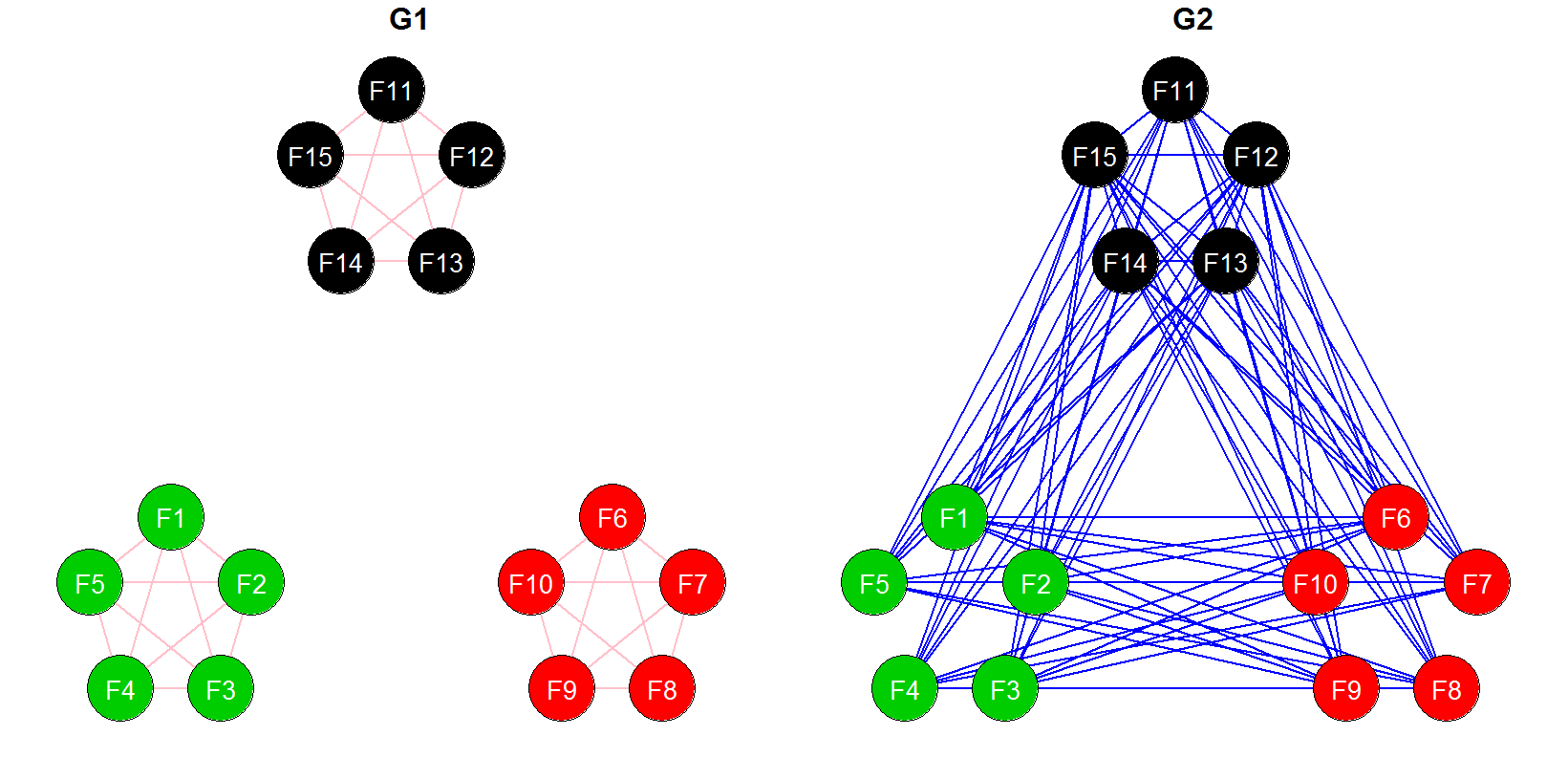
Algorithm command line parameters
- Network 1: edges for intra-module connections, e.g., PPI
- Network 2: edges for inter-module connections, e.g., negative GI
- Initiator: index of the initial solution finder to be used. Options are: 0 for ModMap, 1 for DICER, 2 for hierarchical clustering
- M: minimal module size; e.g., 5
- α: modules link significance threshold; e.g., 0.005
- MTC: binary argument. 1: apply multiple testing correction on the map links; 0 output links with significance <= α
- U: binary argument. 1: run the algorithm on the union of the networks; 0 on the intersection; 0 is preferred for biological data
Command example (on the example data):
java -Xmx2000m -jar ModMap_graphs.jar g1.txt g2.txt 1 5 0.005 1 1The above command will create two text files: one with mapping of nodes to modules,
and another with the map links.
Additional notes
In that case we assume that the executable of that algorithm FP-MBC.exe is in the working directory. You can get this runnable here.
Differential correlation Analysis
Given two (or more) sets of expression profiles labeled generically by the classes 'case' and 'control', ModMap seeks a module-map in which each pair of linked modules correspond to two sets of genes such that each set is highly co-expressed across all profiles, but the correlation between the sets is showing significantly higher co-expression in one class than in the other.
Input data
The input to the algorithm are two files: one is a gene expression matrix, and the second contains the definition of the classes. The gene expression matrix format is a tab-delimited text file in which rows correspond to genes and columns to samples. The classes file is a tab delimited table in which each row represents a class. The first column is the class name, the second is the index of the first sample of the class and the third column is the index of the last sample of the class (note that we assume that the columns of the gene expression matrix are ordered by the class of the samples).Example of input files can be found here
These files represent gene expression data of cases and controls, in which the following linked modules are present:
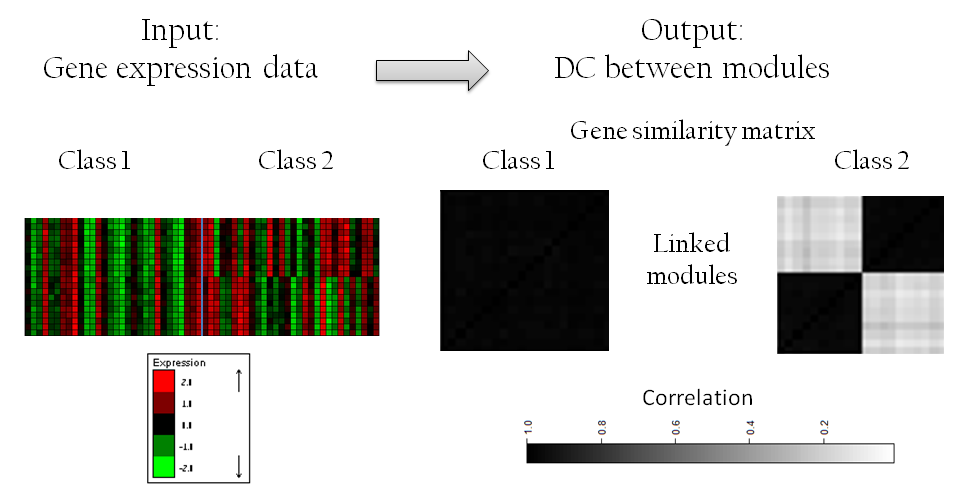
Algorithm command line parameters
- Gene expression data matrix
- Classes file
- class of interest index
- Initiator: index of the initial solution finder to be used. Options are: 0 for ModMap, 1 for DICER, 2 for hierarchical clustering
- M: minimal module size; e.g., 5
- α: modules link significance threshold; e.g., 0.0001
- MTC: binary argument. 1: apply multiple testing correction on the map links; 0 output links with significance <= α
Command example (on the example data):
java -Xmx2000m -jar ModMap_DC.jar ge_mat.txt classes_file.txt 0 1 5 0.0001 1The above command will create two text files: one with mapping of nodes to modules,
and another with the map links (i.e., a single link between the modules above).
Additional notes
In that case we assume that the executable of that algorithm FP-MBC.exe is in the working directory. You can get this runnable here
References
[1] Jinyan Li, Haiquan Li, Donny Soh, Limsoon Wong.A Correspondence Between Maximal Complete Bipartite Subgraphs and Closed Patterns.
Proceedings of 9th European Conference on Principles and Practice of Knowledge Discovery in Databases , pages 146--156, Porto, Portugal, October 2005.
Citing ModMap
ModMap can be cited as follows:Constructing module maps for integrated analysis of heterogeneous biological networks,
David Amar, and Ron Shamir. Nucleic Acids Research doi:10.1093/nar/gku102, 2014.
Get in touch
In case of any questions or suggestions please feel free to contact David Amar.