 |
DICER - Differential Correlation in Expression for meta-module Recovery
DICER is a tool for advanced analysis of differential co-expression (DC) case-control gene expression data.
Given two (or more) sets of expression profiles labeled generically by the classes 'case' and 'control', it seeks two types of groups of genes:
- A set of genes showing significantly higher co-expression in one class than in the other (a DC cluster)
- Two sets of genes such that each set is highly co-expressed across all profiles, but the correlation between the sets is showing
significantly higher co-expression in one class than in the other (a meta-module).
DICER was developed by David Amar with Hershel Safer in Ron Shamir's Computational Genomics group at Tel Aviv University.
|
Differential co-expression signals
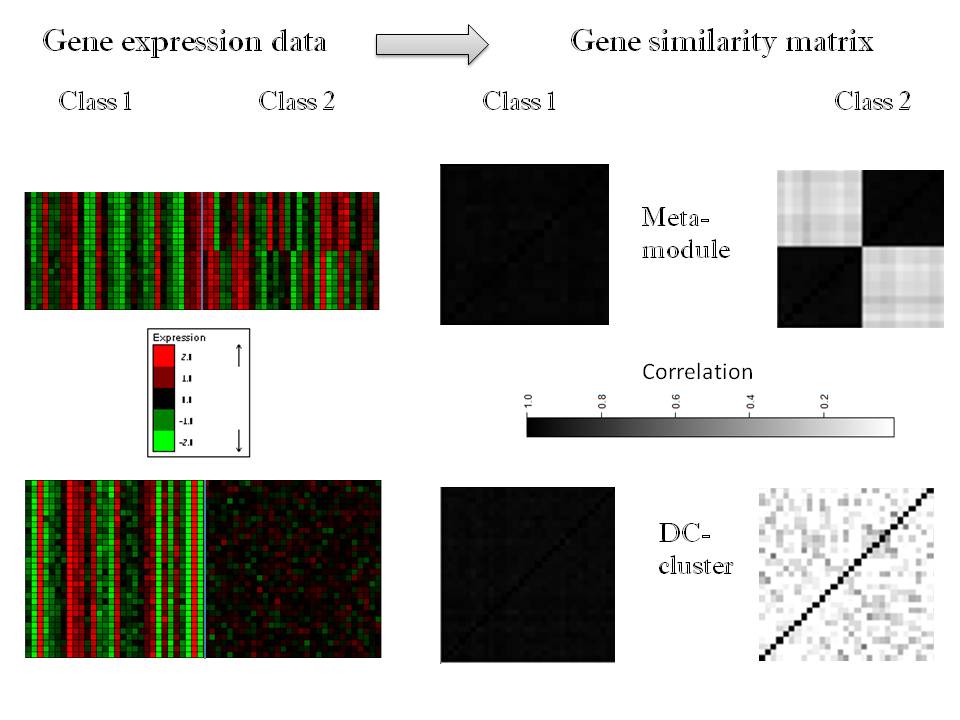 |
Examples of differential co-expression patterns that DICER looks for.
For each class and a gene set the gene expression data and the similarity matrix are shown. |
Get the software
Java executable distribution
Sample datasets
DICER code API
Download the source code from here
To work with the code, download this jar (from EXPANDER)
This distribution is our officially supported executable for DICER.
This binary is completely self-contained and should work out of the box without any issues.
The package includes a small test dataset.
The software is freely available under the GNU Lesser General Public License, version 3, or any later version at your choice.
DICER is a research tool, still in the development stage.
Hence, it is not presented as error-free, accurate, complete, useful, suitable for any specific application or free from any infringement of any rights.
The Software is licensed AS IS, entirely at the user's own risk.
How to use it
java -jar dicer.jar <expression matrix> <classes file> <index of the class of interest><output files prefix><Optional: out=1 to print the graphs><Optional: K=value (default is 2)>
Detailed description of parameters here or in the API.
Examples (Windows):
java -jar dicer.jar AD\top3000.txt AD\classesFile.txt 0 ad_results.txt
java -jar dicer.jar AD\top3000.txt AD\classesFile.txt 0 ad_results.txt out=1
java -jar dicer.jar AD\top3000.txt AD\classesFile.txt 0 ad_results.txt out=1 K=1
Interpreting the output
All output files prefixes are the name of the output path specified by the user.
DICER creates the following files, according to the suffixes:
- _Modules.txt - a flat file with the gene to modules assignments.
in this file, up or down specifies from which DICER analysis the modules where detected.
- The output file (the output path indicated by the user) contains all gene to meta-module or DC cluster mappings.
- _metaGraph.sif - a SIF file that represents the DC map among the modules.
- _metaGraph.txt - a file with the edge scores for _metaGraph.sif
Citing DICER
DICER can be cited as follows:
Dissection of regulatory networks that are altered in disease via differential co-expression, PLoS ComBio 2013
David Amar, Hershel Safer, Ron Shamir.
Improved algorithms for analyzing a pair of networks
We developed an improved algorithm, ModMap, for detecting module maps in a pair of networks.
Get in touch
In case of any questions or suggestions please feel free to contact David Amar.

