|
|
|
|
Overview
|
|
Download and install
The Amadeus software, as well as files with regulatory sequences of various
organisms, can be downloaded from the Amadeus
download page.
After you download the zip file, please extract its contents and follow
the installation instructions in the "README.txt" file.
Note that Amadeus requires Java version 1.5
(get Java here).
How to run Amadues
Following are step-by-step instructions on how to run Amadeus and analyze its output.
 |
|
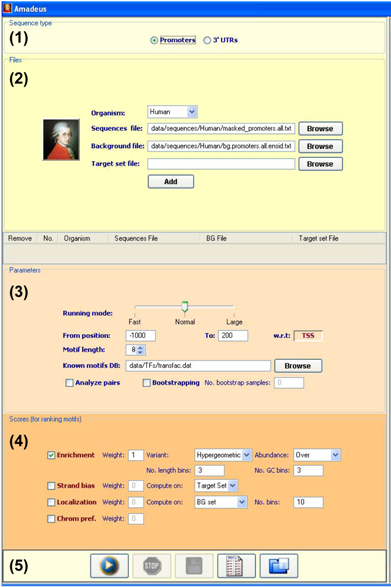
Screenshot of the Amadeus input panel |
- Choose the sequence type
Choose the type of sequences you would like to analyze:
promoters (to analyze both strands) or 3' UTRs (single strand).
- Set files
Choose the organism and assign its sequences and background (BG) & target sets.
When you choose an organism, the default sequences and BG file are those
that you can download from the
download page.
Use the "Browse" button to select different files.
Note that the sequences file should be in fasta format,
in which the header of each sequence contains its name (>geneName),
and the TSS of each gene is assumed to be located at the end of the sequence.
Repetitive elements and other sequences you'd like to ignore in the analysis,
such as protein-coding sequences, should be masked out with N's.
The "BG file" field can be left empty when you want all the genes in the sequences file
to be used as the BG set. Otherwise, the supplied BG file should contain a list of
gene names, each one in a separate line.
The same format applies to the target set, which should be a subset of the BG set.
Typically, the BG set is much larger than the target set, e.g., the BG set are all
the genes represented on the microarray, and the target set is a group of
co-expressed genes.
When using our sequences files, the gene names must be either Ensembl or
Entrez gene ids.
Important notes:
(1) We recommend that the number of BG sequences be at least 5 times larger
than the number of target-set sequences. In general, the larger the BG set is,
the more chances are that Amadeus would recover the regulatory motifs.
The size of the BG set is limited to roughly 65,000 sequences.
Each sequence may be up to 16,000 bases long.
(2) The BG set should be of the same nature as the target set. For example,
if the target set is comprised of genes that were up-regulated in a gene
expression experiment, then the BG set should contain all the genes on the
expression chip.
(3) By default, Amadeus reports motifs that occur in at most 25% of the
background sequences; elements that appear more frequently are often not
biologically interesting. There are additional parameters with default values
that cannot be controlled via the graphical user interface.
If you wish to modify the default parameters of Amadeus, or execute the
program in batch (command-line) mode, please
contact us.
After all the fields have been assigned, press the "Add" button.
Repeat these steps in order to add more organisms/target sets for the analysis.
- Set general parameters
Running mode: choose between faster execution and more comprehensive analysis.
From/to position: determine the range of sequences that will be scanned for the motifs.
Known motifs DB: a file that contains PWMs in Transfac format. The motifs discovered by Amadeus are compared to the PWMs in the file, and similarities are reported.
By default, Amadeus uses Transfac and miRBase for comparison in promoter and 3'-UTR analysis, respectively.
Analyze pairs: choose whether to perform motif-pair analysis, which searches for co-occurring motifs.
Bootstrapping: in order to empirically evaluate the significance of the motif scores, the bootstrap process repeats the entire analysis on <No. bootstrap samples> random gene sets of the same size as that of the supplied target set. Note that this process can take several hours.
- Select score(s) for ranking the motifs
Each motif considered by Amadeus is evaluated using one or more scores.
When several scores are chosen, you may assign them different weights.
All scores are combined into a single p-value.
Enrichment: evaluates the over-representation of the motif in the target set
w.r.t. the BG set. Choose one of the variants: "hypergeometric" or "binned".
The latter accounts for length and GC biases (e.g., when the target set sequences
are more GC-rich than the BG sequences), as described in the paper.
Strand bias, localization, chromosomal preference: evaluate global spatial features
of the motif, namely, whether it's distributed un-evenly between the strands, along the
sequences, or among the chromosomes. These scores may also be used when the target set
is the entire BG set, i.e., when searching for motifs without using a subset of
co-expressed genes.
- Start the analysis
Click the "Run" button to start the analysis.
Other buttons in the bottom panel are: Stop run,
Save textual output to file, Save parameters to file, Load parameters from file.
- Output of Amadeus
Amadeus has both a textual and graphical output.
At the top of the textual "Output" tab, Amadeus reports general statistics on the
supplied input, e.g, the number of BG/target-set sequences, their average length and
their base frequencies. Check these stats to verify that your input was read correctly.
Once the analysis is completed, Amadeus shows the discovered motifs in the
graphical "Results" tab (see figure below).
If pairs analysis was chosen, the results are shown in an additional tab.
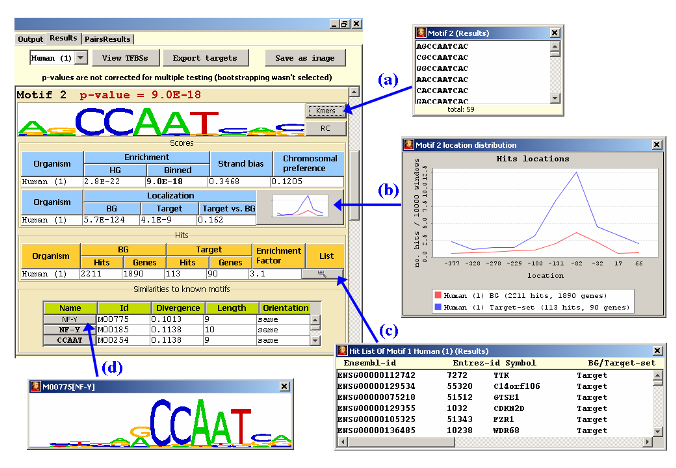
For each discovered motif, Amadeus reports its p-value (and fixed p-value,
in case bootstrapping was executed), its graphical logo, the scores it attained (all scores
are shown; those used for computing the p-value are marked in bold face),
statistics on the number of hits and targets, and a list of similar known motifs
from Transfac/miRBase ("Divergence" closer to 0 means higher similarity).
Additional information is presented in several pop-up screens (see figure):
(a) The list of k-mers that comprise the motif (i.e., pass the PWM cutoff).
(b) A histogram of the locations of the motif's hits in the BG (red) and
target set (blue) sequences. Location 0 is the TSS (or, in 3' UTR analysis,
the 3' end of the sequence).
(c) A list of genes whose promoters/3'-UTRs contain a hit of the reported motif.
This list can be exported for further analysis.
(d) The logo of the chosen PWM from Transfac.
|
|



