| Home |
|
Amadeus home |
| Overview |
|
Amadeus software overview |
| Download |
|
Download Amadeus and the compendium |
| Supplementary data |
|
Supplementary data for the paper |
| Contact us |
|
|
| Home | ||
|
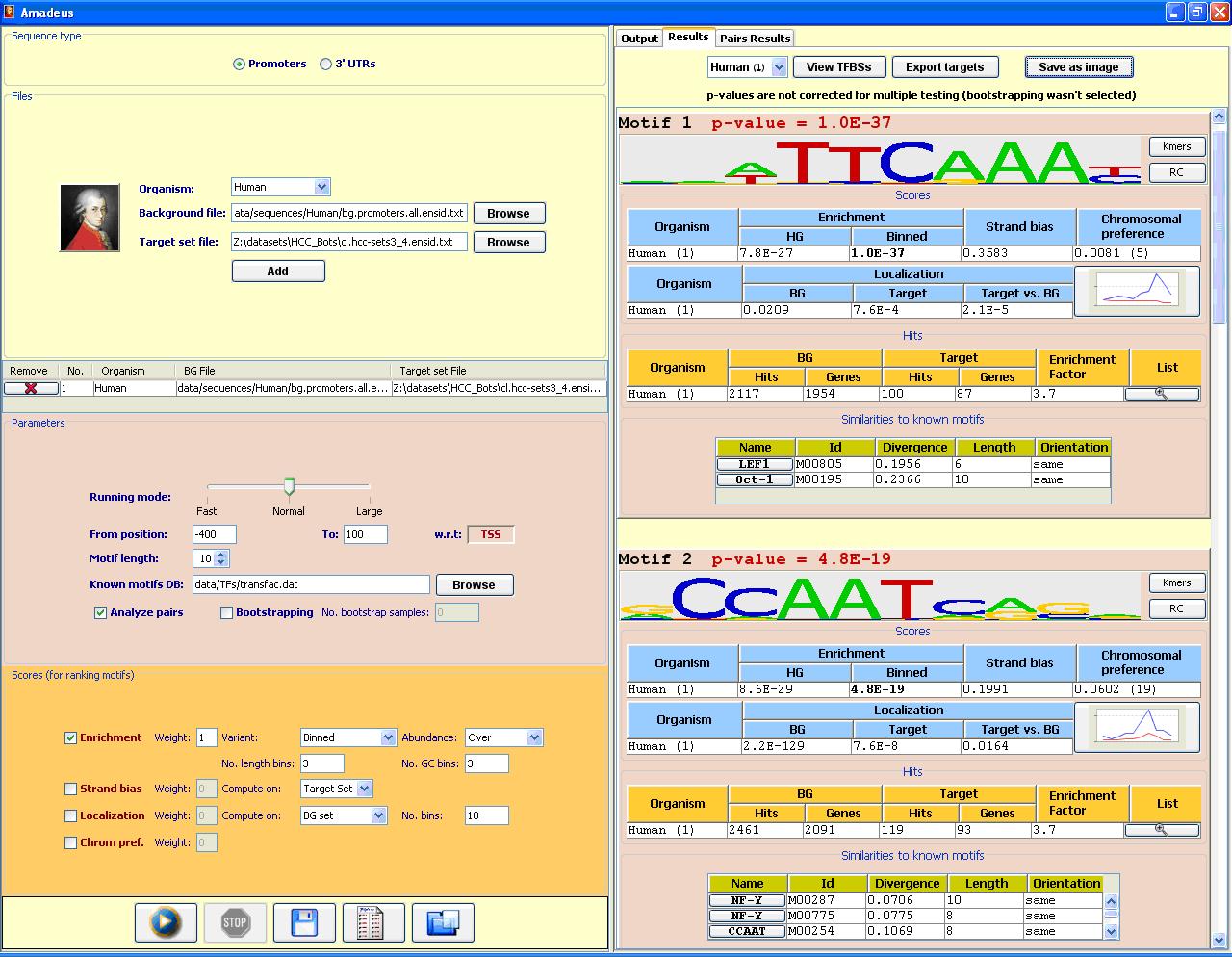
The Amadeus motif discovery platformAmadeus is a user-friendly software platform for genome-scale detection of known and novel motifs (recurring patterns) in DNA sequences, applicable to a wide range of motif discovery tasks. Amadeus outperforms extant tools and sets a new standard for motif discovery software in terms of accuracy, running time, range of application, wealth of output information and ease of use. PBM analysis: Amadeus can be used to identify binding site motifs from Protein Binding Microarray (PBM) data. A PBM dataset of a specific TF includes its measured binding intensities for each probe sequence covering together all possible 10-mers. In a blind competition (DREAM5 Challenge, Sep. '10) Amadeus ranked first (tied with one other group) in identifying the binding site motifs of 66 TF datasets. Running time is a few seconds per dataset. Note: Amadeus is now integrated with Allegro, our new motif-finding algorithm for discovering transcriptional modules in gene expression datasets. Revisions are made only to the combined Amadeus/Allegro version. Visit the Allegro web-site for more details and for the latest software and data.
Amadeus is described in our paper:
Amadeus-PBM is described in our paper:
This web-site contains the following:
|