|
Download and install
How to run AmadeusPBM/Allegro/Amadues
Important notes
Why "Allegro"?
Download and install
- Download the software from the
download page.
After you download the zip file, please extract its contents to a folder
of your choice, e.g., D:\MyTools. This should create a directory named
AmadeusPBM_v1.0 in the above folder.
- Download regulatory sequences from the
download page.
Save the sequences zip file in the Allegro directory, e.g., under
D:\MyTools\AmadeusPBM_v1.0 in the previous example.
Then, extract the zip file and make sure the sequences files reside in the
data\sequences\[org] folder, where [org] is the name of the organism.
For example, if you download human promoters, you should verify that the
directory D:\MyTools\AmadeusPBM_v1.0\data\sequences\Human contains a
file called "masked_promoters.all.txt".
- Further instructions, hardware/software requirements and notes
are detailed in the README.txt file supplied with the installation.
Note that Allegro requires Java version 1.5 or later
(get Java here),
and at least 1GB RAM memory.
Allegro should run properly on Windows (2000/XP/Vista) and
Linux (Debian). It was not tested on Mac OS.
How to run Allegro/Amadues
Following are step-by-step instructions on how to run Allegro and Amadeus,
and analyze their output.
 |
|
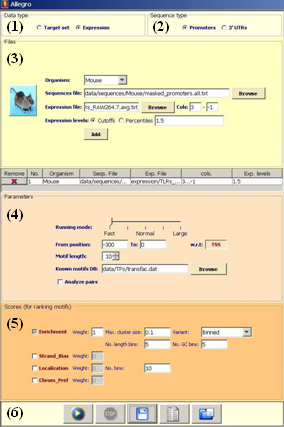
Screenshot of the Allegro input panel |
- Launch the applcation
Execute/double-click the "run.bat" file in the Allegro folder.
If you wish to analyze a very large expression file or multiple datasets,
you may need to allocate more memory for Java. To do so, either execute
the "run_1.3G_mem.bat" file, or edit the "run.bat" file and change the
value of "-Xmx". Note that you will need at least 2GB RAM memory for such
large executions.
- Choose the data type
Select "PBM" if you are analyzing PBM dataset. This will run the
AmadeusPBM algorithm. Follow the rest of the instructions on this page.
Select "Expression" if you are analyzing a gene expression dataset (or similar
types of data that assign one or more values to each gene). This will run the
Allegro algorithm. Follow the rest of the instructions on this page.
Select "Target set" if you would like to execute the Amadeus algorithm in order
to:
(a) discover motifs that are over-represented in a supplied list of genes (or
several such lists); or (b) find motifs with global spatial features
(i.e., motifs that appear non-uniformly along the promoters, between the two
strands, or among the chromosomes). For more information on the input of Amadeus,
please read the
Amadeus overview page.
- Choose the sequence type (if "Expression" or "Target set")
Choose the type of sequences you would like to analyze:
promoters (to analyze both strands) or 3' UTRs (single strand).
- Set files
Choose the organism and its sequences & expression files.
PBM file (in case of "PBM"):
Use the "Browse" button to select a file.
Note that the PBM file should be in deBruijn format (from UniProbe database),
in which each sequence's prefix contains 36 informative base pairs and
a tab delimited number as a measur of the binding.
Sequences (if "Expression" or "Target set"):
When you choose an organism, the default sequences file is set to
the file you can download from the
download page.
Use the "Browse" button to select a different file.
Note that the sequence file should be in fasta format,
in which the header of each sequence contains its name (>geneName),
and the TSS of each gene is assumed to be located at the end of the sequence.
Repetitive elements and other sequences you'd like to ignore in the analysis,
such as protein-coding sequences, should be masked out with N's.
The sequence file may contain up to 65,000 sequences, each up to 16,000 bases long.
Expression (if "Expression"):
Use the "Browse" button to select the expression data file.
The file should be tab-delimited, and contain one line per gene.
Each line should contain the id of the gene in the first column,
and the expression values in the other columns. Specify the range
of columns you'd like to include in the analysis using the "Cols" boxes.
For example, if you'd like to ignore the 2nd column in the expression
file, use "Cols" 3 to -1 ("-1" stands for the last column).
The first line in the expression file should specify the names (titles) of the
experimental conditions.
If the second line starts with ">SERIES", then it should indicate
the series name of each condition. This feature is optional, and is intended
for presentation purposes only - it has no effect on the algorithm's results.
The rest of the lines in the expression file should contain the expression
values, as mentioned above.
An example of the expression file format can be found
here.
See also the file "TLRs_RAW264.7.avg.txt" in the "expression" directory
of the installation.
Important notes:
(1) Allegro does not perform any type of normalization to the data. Thus, it is
up to the user to pre-process the data properly, as explained
here.
(2) The gene ids should match those in the sequences file. If you are using our
sequences files, the gene names should be either Ensembl or Entrez gene ids.
Genes in the expression file that aren't found in the sequences file, or vice
versa, are ignored.
Expression levels:
As described in the paper, Allegro uses discrete expression levels in its
CWM exression model. The discretization is performed either using fixed cutoffs
or using percentiles. If you choose "Cutoffs", you should specify the lower bound
of each expression level except the lowest one. E.g., if the cutoffs
are "1.5, -1", Allegro will use three levels: 1.5 and up, between -1 and 1.5,
and less than -1. If you specify "Percentiles" as "10,90", for example, then
Allegro will use two expression levels in each condition: the top 10%, and
the rest 90%.
Note that the cutoffs/percentiles apply to all the conditions in the expression file.
PBM parameters:
As described in the paper, AmadeusPBM uses 3 parameters: scoring scheme, k and
number of k-mers. Each is configurable through the GUI. The scoring scheme is
either average, median or WMW. k should be set between 7 to 10 (=k-mer length). The default
number of top k-mers value is 1000, but you may want to experiment with other values.
After all the fields have been assigned, press the "Add" button.
Repeat these steps in order to add more organisms/expression files to the analysis.
- Set general parameters
Running mode: choose between faster execution and more
comprehensive analysis.
From/to position: determine the range of sequences that will be
scanned for the motifs.
Motif length: the length of the motifs to search for. We recommend
values of 8-10. If "PBM" you should set it to be at most the chosen k.
Known motifs DB: a file that contains PWMs (matrices) of known motifs,
in Transfac format. The motifs discovered by Allegro are compared to the PWMs
in the file, and similarities are reported.
By default, Allegro uses Transfac and miRBase for comparison in promoter and
3'-UTR analysis, respectively. Species-specific miRBase files are also
supplied in the installation - see "data/miRNA/README.txt".
Analyze pairs: choose whether to perform motif-pair analysis,
which searches for co-occurring motifs.
Amadeus/Allegro uses additional parameters that have pre-defined
default values and cannot be controlled via the GUI,
as explained here.
- Select score(s) for ranking the motifs
Each motif considered by Allegro is evaluated using one or more scores.
When several scores are chosen, you may assign them different weights.
All scores are combined into a single p-value.
Enrichment: (obligatory) evaluates the over-representation of the motif
in the sequences of genes, which share
the expression profile that Allegro inferred for the motif.
Choose one of the variants: "hypergeometric" or "binned".
The latter accounts for length and GC biases (i.e., when the genes'
expression profiles are correlated to the length and/or GC-content of their
regulatory sequences), as described in the paper.
Strand bias, localization, chromosomal preference: (optional) evaluate global
spatial features of the motif, namely, whether it's distributed un-evenly
between the strands, along the
sequences, or among the chromosomes.
- Start the analysis
Click the "Run" button to start the analysis.
Other buttons in the bottom panel are: Stop run,
Save textual output to file, Save parameters to file, Load parameters from file.
- Output of Allegro
Allegro has both a textual and graphical output.
At the top of the textual "Output" tab, Allegro reports general statistics on the
supplied input, e.g, the number of sequences, their average length and
their base frequencies, the number of conditions read from the expression file,
the distribution of discrete expression levels in each condition, and more.
Check these stats to verify that your input was read correctly.
Once the analysis is completed, Allegro shows the discovered motifs in the
graphical "Results" tab (see figure below).
If pairs analysis was chosen, the results are shown in an additional tab.
As described in the paper, Allegro uses a novel non-parametric model
called CWM (Condition Weight Matrix) to describe the expression profile of a
group of co-regulated genes.
For each candidate motif, Allegro fits a CWM to its putative targets
using a cross-validation-like procedure. The genes whose expression values
match the CWM (above a computed threshold) are called the CWM targets.
In order to ascertain whether
the motif and the CWM are significantly correlated, Allegro computes one of two
enrichment scores, as chosen by the user:
the HG score, or the binned enrichment score.
Allegro utilizes the efficient motif search engine of Amadeus
to enumerate a huge number of candidate motifs and to converge to high-scoring ones.
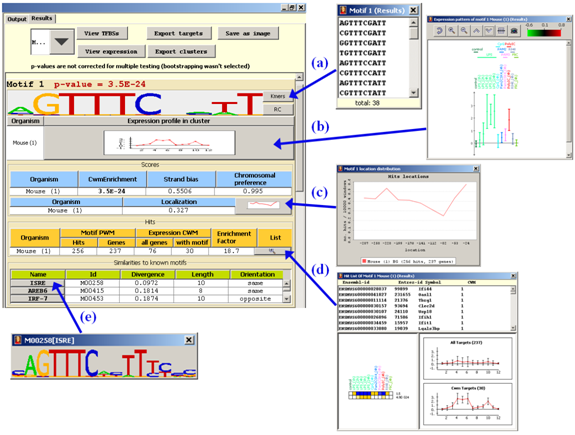
For each discovered motif, Allegro reports its p-value,
its graphical logo, the scores it attained (all scores
are shown; those used for computing the p-value are marked in bold face),
the CWM expression profile fitted to its targets,
statistics on the number of hits and targets, and a list of similar known motifs
from Transfac/miRBase ("Divergence" closer to 0 means higher similarity).
Additional information is presented in several pop-up screens (see figure):
(a) The list of k-mers that comprise the motif (i.e., pass the PWM cutoff).
(b) Expression profile of the motif's targets that are also its CWM targets (i.e.,
genes whose sequence contains a hit of the motif, and whose expression values
match the CWM). Multiple types of views are available (mean expression profile,
per-gene expression matrix).
(c) A histogram of the locations of the motif's hits in the regulatory sequences.
Location 0 is the TSS (or, in 3' UTR analysis, the 3' end of the sequence).
(d) A list of genes whose promoters/3'-UTRs contain a hit of the reported motif.
This list can be exported for further analysis. Also shown are a graphical
representation of the CWM fitted to the motif's targets (bottom left),
and the mean expression profile of: [i] all the targets of the motif (middle right),
[ii] the motif's targets that are also its CWM targets (bottom right).
(e) The logo of the chosen PWM from Transfac.
Important notes
- Default values, batch runs:
The algorithms have many parameters that control the way motifs are
searched and reported. The most important parameters can be set by the user via
the graphical user interface (GUI). However, other parameters are always set with
pre-defined default values. For example, by default, Amadeus reports motifs that
occur in at most 25% of the background sequences; elements that appear more
frequently are often not biologically interesting.
If you wish to modify the default
values of Amadeus/Allegro, or execute the programs in batch (command-line) mode,
you first need to create a parameters file for the execution - you can do this
using the graphical interface (i.e., set all the parameters, run Amadeus/Allegro
to make sure they're ok, and then click the "Save parameters to file" button).
Then, run Amadeus/Allegro by simply adding "file [filename]" to the command
line, where "filename" is the name of the parameters file you saved. For
example:
java -Xmx800m -jar Allegro_v1.0.jar file myParams.txt
Note: On Windows, you need to execute the above line in a command ("DOS")
window (click "Start"->"Run", type "cmd" and hit Enter);
on Linux, execute the above line in a shell (xterm/terminal) window.
Please contact us
in case you need further assistance.
- Pre-processing the expression data:
Before supplying the expression data to Allegro, you should pre-process it
according to the experimental setting and what type of expression profiles you
wish to find. For example, when measuring expression values along a time course,
it is usually recommended to compute the log fold-change in each time point
relative to the first time point. For expression profiles in various tissues or
under different conditions (i.e., when there is no clear base condition),
one could standardize the genes' profiles to mean 0 and SD 1, or compute the
log ratio relative to the average value. Note that you should NOT filter out
genes (e.g., those that did not change in any condition), since Allegro utilizes
as many genes as possible to enhance the power of its statistical tests.
Pre-processing can be performed using any gene expression analysis software,
such as our Expander
platform, or by other means (Excel, R, Perl script, etc.).
Why "Allegro"?
First, the name Allegro is an acronym of "A Log-Likelihood based Engine for
Gene expression Regulatory motifs Over-representation discovery".
Second, in music allegro refers to a quick, lively tempo; the Allegro
software allows a fast (and accurate) analysis of gene expression datasets
in the context of regulatory motif discovery.
Third, Allegro builds upon the Amadeus motif discovery platform, so we
wanted the name of our new method to be from the musical domain as well.
One of the most popular and recognized classical pieces is the first
movement of Wolfgang Amadeus Mozart's Eine kliene Nachtmusik, which is in
sonata allegro form. The background of the Allegro logo shows the
sheet music of this piece.
|



