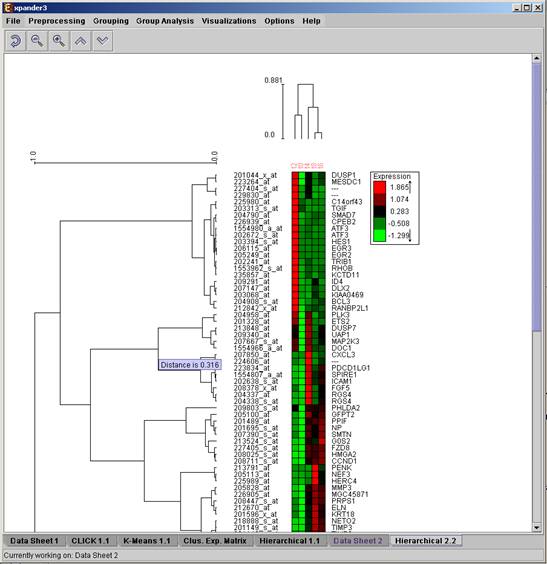
This tool uses the agglomerative algorithm to calculate a dendrogram tree for all expression patterns (probe patterns) and/or profiles (condition profiles). The type of linkage (manner in which the distance between a new node and the rest of the nodes is calculated) used in the algorithm can be set via an input dialog (for details regarding the algorithms refer to the References section). Note that it does not generate a partition of the probes to clusters. The distance measurement used in the algorithm is (1-Pearson Correlation)/2.
To perform hierarchical clustering, select Grouping >> Hierarchical Clustering. Upon selecting this option, a dialog box appears in which the ‘linkage type’ parameter, used in the algorithm can be set. After pressing ‘OK’, the algorithm will be operated both on the probe patterns and on the condition profiles.
The resulting trees are displayed next to an expression matrix so that the probe tree appears vertically on the left and the condition tree appears horizontally above the matrix. The scale next to each tree indicates the range of distance values between vectors corresponding to the leaves. The tool tip indicates the distance value corresponding to the cursor location on the tree.
