| Home |
|
Allegro home |
| Overview |
|
Allegro tool overview |
| Hands On |
|
Amadeus-Allegro tutorial |
| Download |
|
Download Allegro and regulatory sequences |
| Supplementary data |
|
Supplementary data for the paper |
| Contact us |
|
|
| Home | ||
|
Allegro: Discovering DNA motifs from sequence and expression datasets
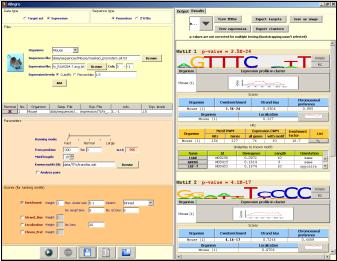
Allegro is a software tool for simultaneous discovery of cis-regulatory motifs and their associated expression profiles. Its input are DNA sequences (typically, promoters or 3' UTRs) and genome-wide expression profiles. Its output is the set of motifs found, and for each motif the set of genes it regulates (its transcriptional module). Allegro is highly efficient and can analyze expression profiles of thousands of genes, measured across dozens of experimental conditions, along with all regulatory sequences in the genome. Allegro has a user-friendly graphical user interface. As of 2009, the Allegro software integrates also the Amadeus motif finding algorithm.
NewsOct. 2017: To accommodate large ChIP-seq datasets, up to 65,000 sequences can now be handled (see overview page).Nov. 2014: Updated files with known regulatory motifs from public databases: Please download the files from the download page. Nov. 2013: Amadeus is now integrated in the Expander software suite for gene expression analysis. In addition to the stand-alone software available till now, it can also be executed as part of the Expander analysis flow. Jun. 2013: The promoters and 3' UTRs available in Allegro have been revised. They are available together with the software from the download page. Jul. 2011: AmadeusPBM is now avilable. This new addition to the software is a tool for extracting binding site motifs from protein binding microarray (PBM) data. You can download the new AmadeusPBM from the download page. Apr. 2011: Updated micro-RNA seed motifs from miRBase 12.0 to miRBase 16.0. Please download the new files from the download page. Sep. 2010: The DREAM5 Challenge - Amadeus ranked first in the DREAM5 Challenge for identifying the binding site motifs of 66 PBM (Protein Binding Microarray) datasets (see below).
Motif discovery tasksThe Allegro software package also includes our recently published Amadeus motif discovery platform. Thus, the integrated software supports the following motif finding tasks:
PBM analysis: Amadeus can also be used to identify binding site motifs from Protein Binding Microarray (PBM) data. A PBM dataset of a specific TF includes its measured binding intensities for each probe sequence covering together all possible 10-mers. In a blind competition (DREAM5 Challenge, Sep. '10) Amadeus ranked first (tied with one other group) in identifying the binding site motifs of 66 TF datasets. Running time is a few seconds per dataset.
PublicationsAllegro, Amadeus and AmadeusPBM are described in our papers:
|