| sce | p-val = 114.456, 178 hits in 112 genes |  | p-val = 15.1431, 46 hits in 38 genes |  |
| paradoxus | p-val = 44.4381, 70 hits in 46 genes |  | p-val = 14.8037, 57 hits in 31 genes |  |
| kudri | p-val = 53.9531, 93 hits in 57 genes |  | p-val = 20.5318, 67 hits in 46 genes |  | p-val = 7.06195, 38 hits in 36 genes |  |
| mik | p-val = 51.2573, 76 hits in 48 genes |  | p-val = 13.1844, 49 hits in 40 genes |  | p-val = 9.46266, 32 hits in 29 genes |  | p-val = 7.85091, 41 hits in 36 genes |  |
| bayanus | p-val = 49.9982, 87 hits in 54 genes |  | p-val = 16.3118, 68 hits in 40 genes |  | p-val = 8.0054, 40 hits in 36 genes |  |
| cgal | p-val = 29.2958, 105 hits in 45 genes |  | p-val = 17.7804, 23 hits in 21 genes |  | p-val = 16.0612, 42 hits in 29 genes |  | p-val = 8.55818, 45 hits in 34 genes |  |
| castelli | p-val = 58.7922, 85 hits in 53 genes |  | p-val = 38.2149, 53 hits in 45 genes |  | p-val = 31.5228, 80 hits in 40 genes |  | p-val = 8.65079, 41 hits in 31 genes |  | p-val = 8.27505, 25 hits in 24 genes |  | p-val = 7.50176, 33 hits in 31 genes |  |
| klac | p-val = 32.84, 46 hits in 40 genes |  | p-val = 22.2416, 48 hits in 35 genes |  | p-val = 15.5654, 23 hits in 22 genes |  | p-val = 12.522, 30 hits in 17 genes |  | p-val = 7.31282, 18 hits in 17 genes |  |
| klu | p-val = 21.9569, 44 hits in 30 genes |  | p-val = 20.9881, 59 hits in 32 genes |  | p-val = 10.844, 30 hits in 29 genes |  | p-val = 8.58251, 41 hits in 31 genes |  | p-val = 7.77608, 31 hits in 24 genes |  |
| kwal | p-val = 40.0412, 99 hits in 31 genes |  | p-val = 35.4911, 46 hits in 34 genes |  | p-val = 29.6101, 32 hits in 30 genes |  | p-val = 19.608, 35 hits in 27 genes |  | p-val = 11.6895, 34 hits in 29 genes |  |
| agos | p-val = 77.7546, 122 hits in 64 genes |  | p-val = 36.3141, 69 hits in 52 genes |  | p-val = 21.8467, 72 hits in 32 genes |  | p-val = 9.32758, 32 hits in 28 genes |  |
| dhan | p-val = 35.0269, 60 hits in 46 genes | 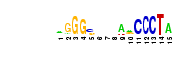 | p-val = 31.0165, 116 hits in 66 genes |  | p-val = 28.678, 49 hits in 41 genes |  | p-val = 15.2369, 60 hits in 46 genes |  | p-val = 14.4213, 54 hits in 44 genes |  | p-val = 10.107, 51 hits in 39 genes |  | p-val = 9.91683, 45 hits in 35 genes |  | p-val = 8.09796, 48 hits in 38 genes |  | p-val = 8.06223, 50 hits in 39 genes |  | p-val = 7.84089, 49 hits in 39 genes |  | p-val = 7.73513, 49 hits in 38 genes |  | p-val = 7.40888, 44 hits in 36 genes |  |
| cab | p-val = 34.3928, 35 hits in 30 genes |  | p-val = 14.9279, 39 hits in 32 genes |  | p-val = 11.0017, 22 hits in 17 genes |  | p-val = 8.07284, 10 hits in 10 genes |  | p-val = 7.27583, 32 hits in 21 genes |  |
| ylip | p-val = 39.2681, 63 hits in 51 genes |  | p-val = 30.2554, 97 hits in 53 genes |  | p-val = 23.0991, 60 hits in 47 genes |  | p-val = 21.0588, 49 hits in 40 genes |  | p-val = 7.67671, 44 hits in 31 genes |  |
| ncr | p-val = 25.239, 60 hits in 46 genes |  | p-val = 21.9773, 73 hits in 46 genes |  | p-val = 14.798, 58 hits in 42 genes |  | p-val = 10.0573, 48 hits in 39 genes |  | p-val = 8.73265, 53 hits in 39 genes |  | p-val = 8.44465, 39 hits in 32 genes |  | p-val = 7.63726, 38 hits in 33 genes |  |
| ani | p-val = 24.9884, 71 hits in 48 genes |  | p-val = 21.7595, 57 hits in 49 genes |  | p-val = 21.3575, 51 hits in 39 genes |  | p-val = 15.1093, 36 hits in 33 genes |  | p-val = 7.26372, 57 hits in 32 genes |  |
| pombe | p-val = 91.3836, 93 hits in 73 genes |  | p-val = 43.0452, 54 hits in 44 genes |  |





























































































































































