

TWIGS - Three-Way module Inference via Gibbs Sampling
TWIGS is a tool for advanced analysis of three-way data (e.g., patient-gene-time in gene expression or subject-voxel-time in fMRI). TWIGS identifies both core modules that appear in multiple patients and patient-specific augmentations of these core modules that contain additional genes.Toy Example
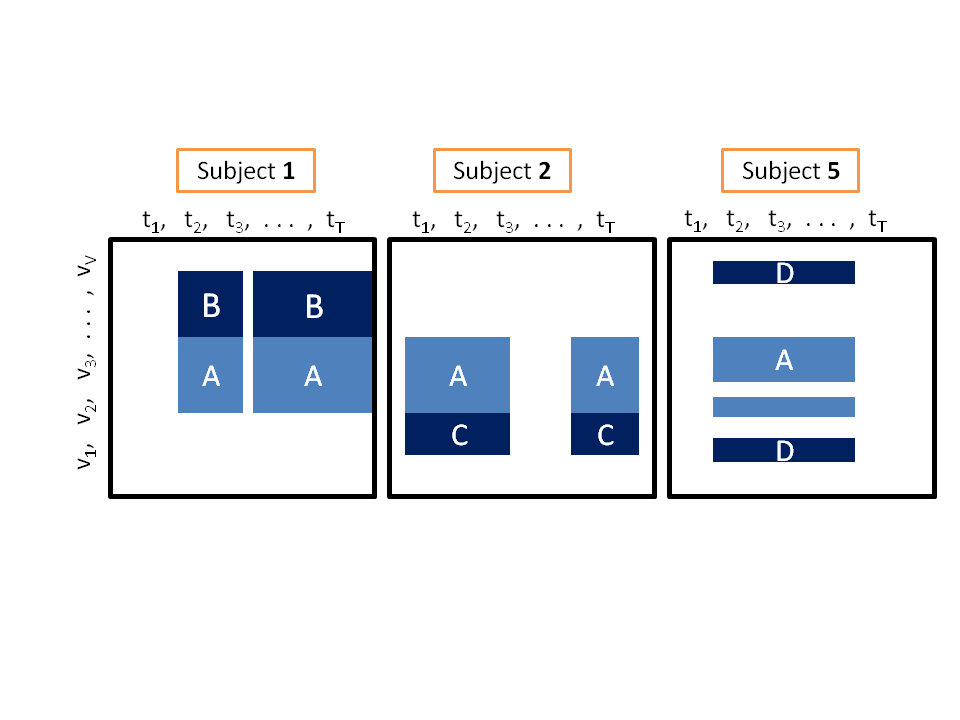 |
The three-way data example above has a row for each gene. Each column represents a specific time point in the data of a subject. The area A represents a core module: (1) it has a set of rows that are likely to be active together across a set of subjects, and (2) it has a set of active time points in each covered subject, and these sets differ between subjects The other areas represent subject-specific augmentations of the core module. |
Get the software
TWIGS is a research tool, still in the development stage.Hence, it is not presented as error-free, accurate, complete, useful, suitable for any specific application or free from any infringement of any rights. The Software is licensed AS IS, entirely at the user's own risk.
| Name (link) | Description |
|---|---|
| Gibbs sampling implementation | R implementation of TWIGS |
| Helper functions | Auxiliary functions, e.g., implementation of the simulations |
| Run commands | R commands for producing the TWIGS results on the real data (fMRI and gene expression) |
Get the datasets
Citing TWIGS
TWIGS can be cited as follows:A hierarchical Bayesian model for flexible module discovery in three-way time series data,
David Amar, Daniel Yekutieli, Adi Maron-Katz, Talma Hendler, and Ron Shamir. To appear in ISMB 2015.
Get in touch
For questions or suggestions please contact David Amar.