Yeast Regulatory Selection
Network
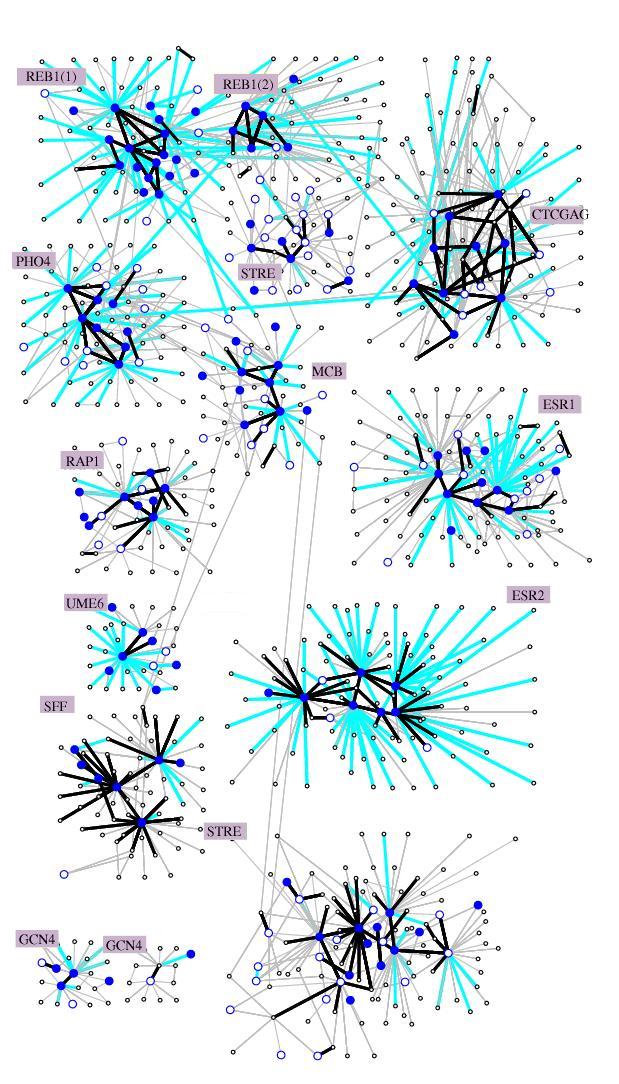
Partial view of the selection network. Motifs are represented as circles;
motifs that have conservation rate > 0.2 at 2 SD are shown by large circles.
Motifs with conservation rate > 0.2 at 3 SD are shown as filled circles.
Substitutions are shown as color coded arcs. Black: high substitution rate
(r>0 and r>-0.5 at 2 SD); cyan: mnegative rates (r<-0.5 at 2 SD).
A family in the selection network is a collection of motifs that are interconnected
via high or neutral substitution rate arcs. Families are separated by low
rate substitution arcs. We annotated each family
by a consensus motif, and by the names of the transcription factors that
bind to motifs in the family (if such are known). Numerous known transcription
factors are identified as families of motifs, which can be further analyzed
for intricate intra-family relations.
Clicking a large circle open the M
otif Specific Substitution M
atrix of the corresponding motif.
Database revision : yeast_stricto
this is an automaticly generated PREVO report
Computational Genomics Lab, Tel-Aviv uniresity
