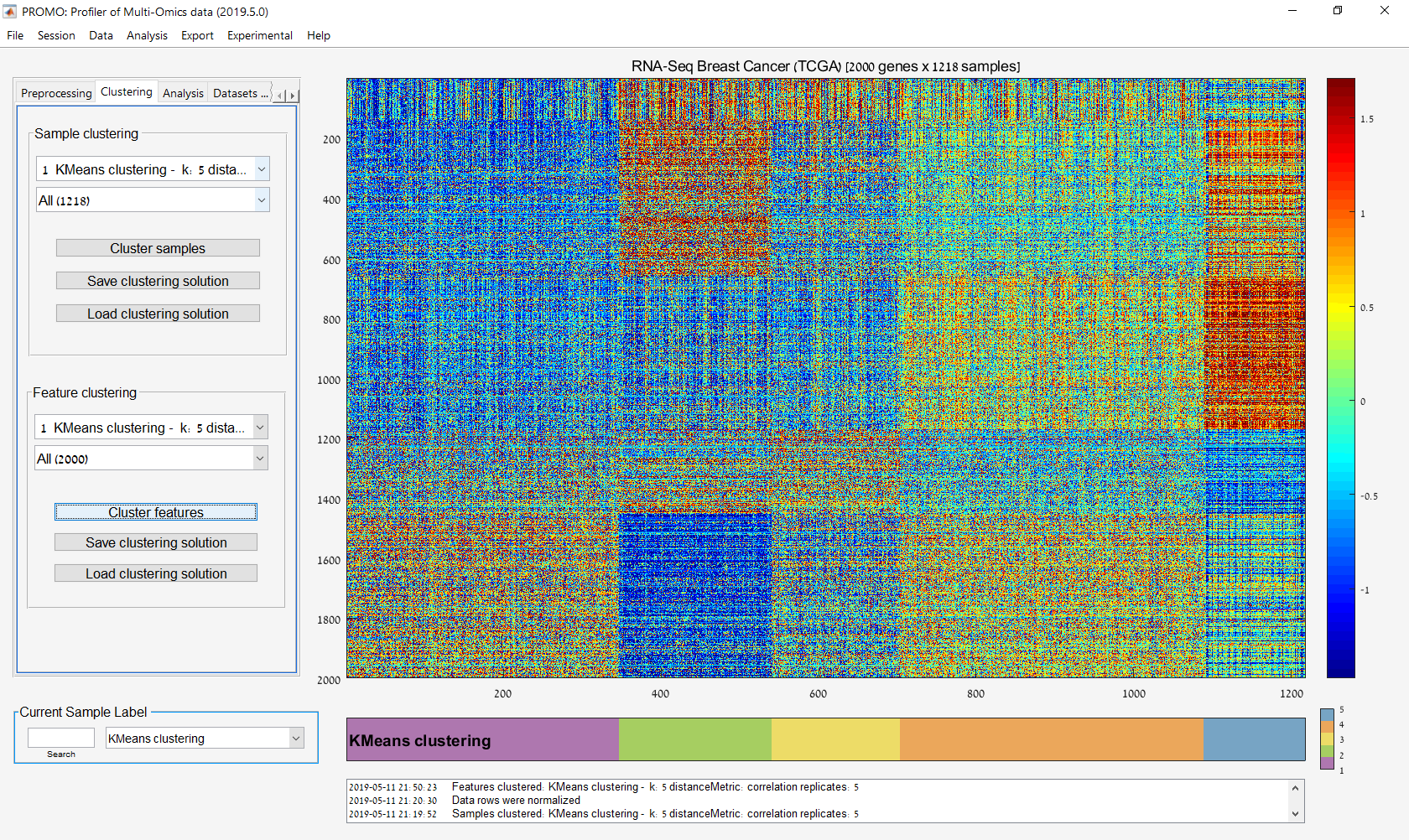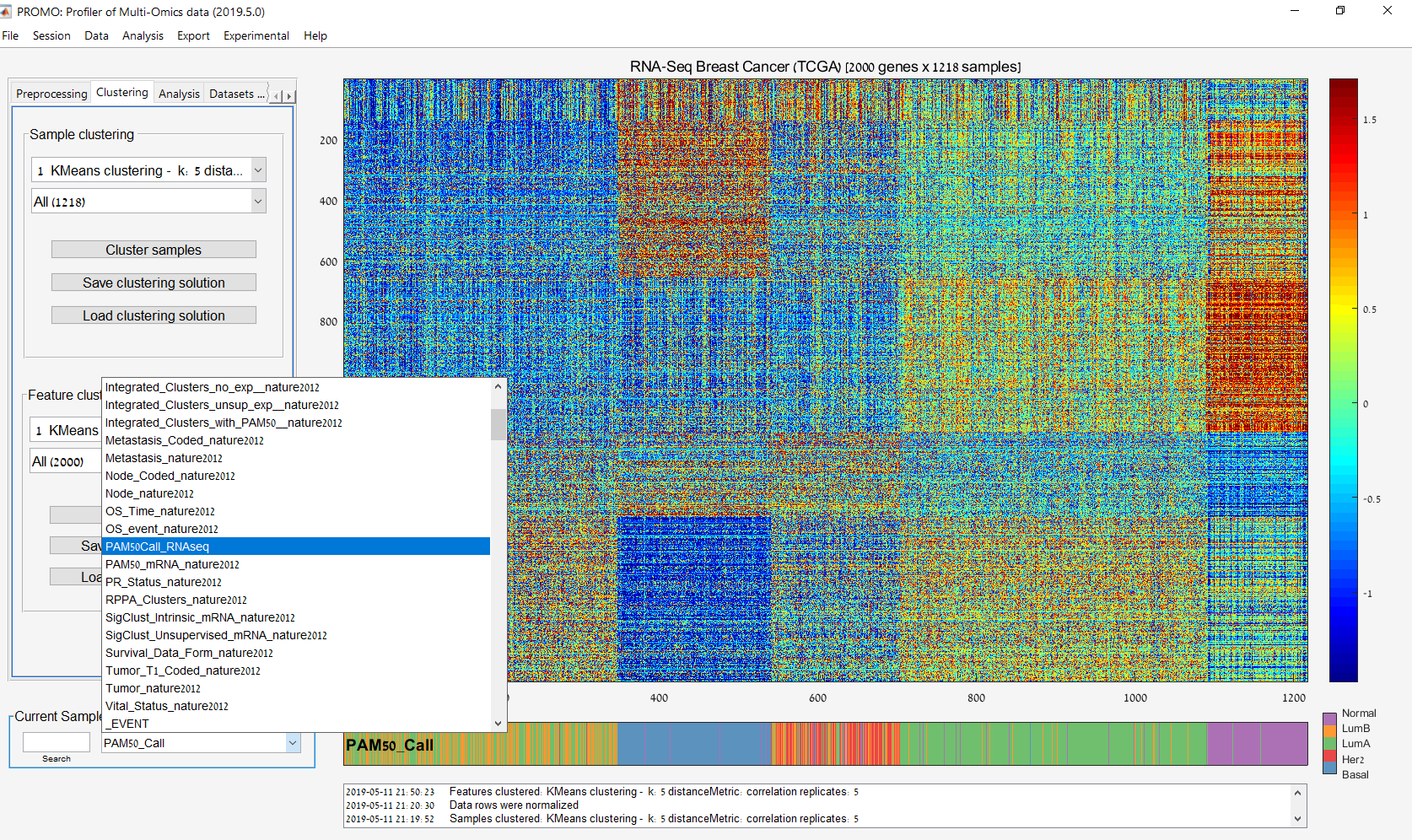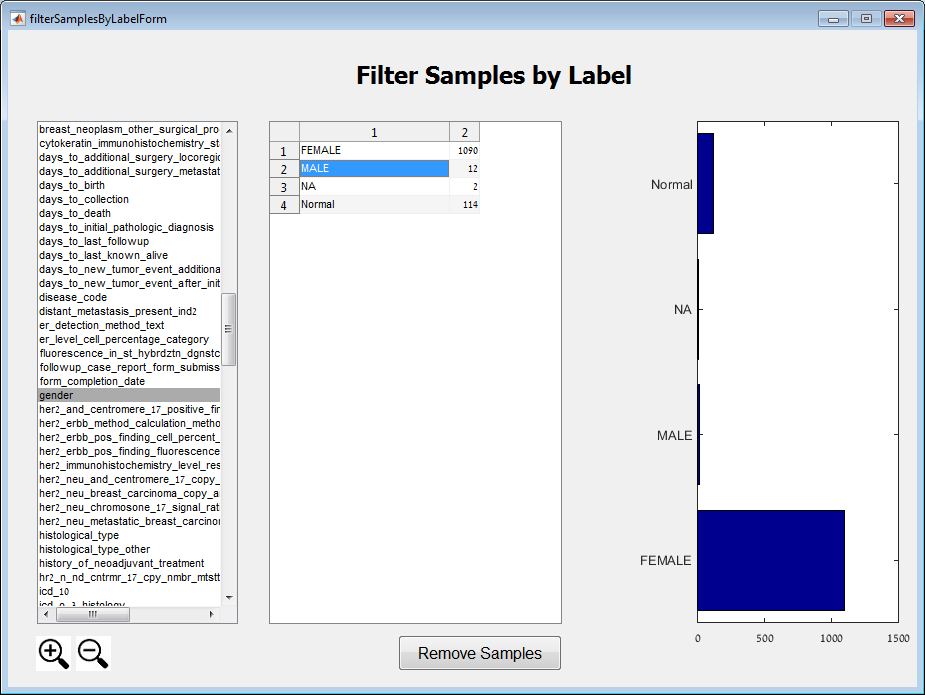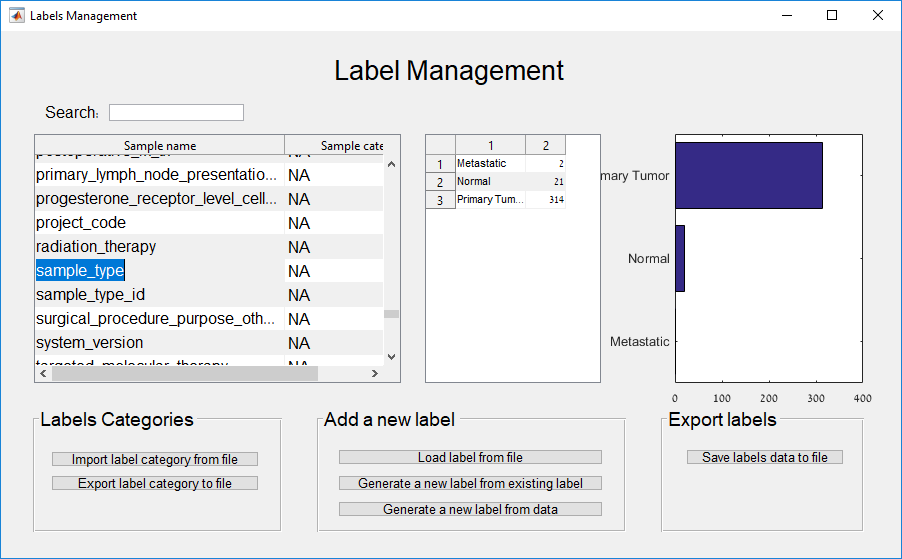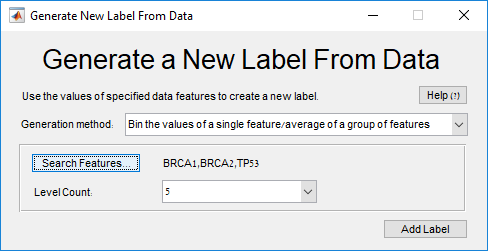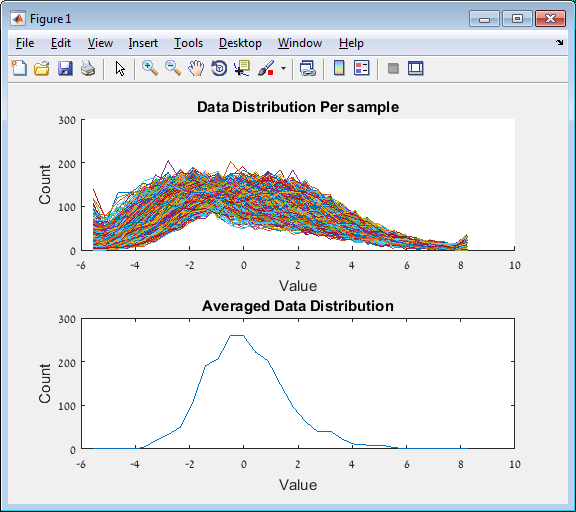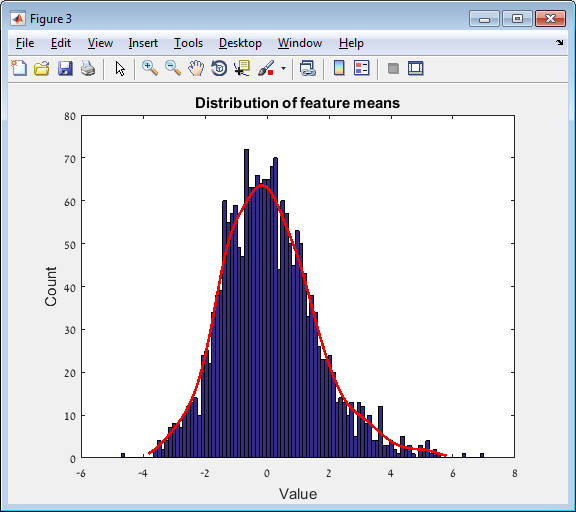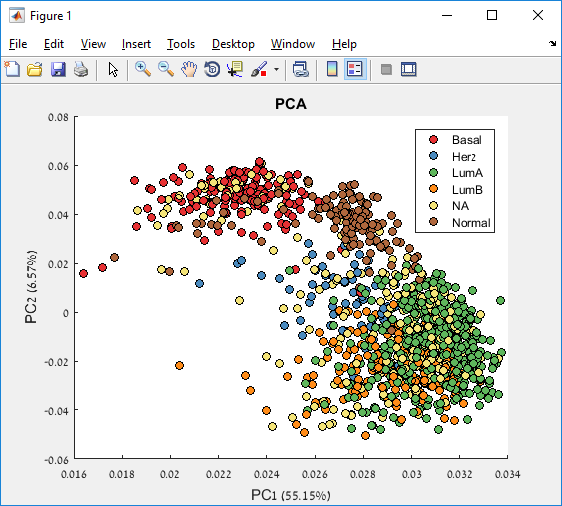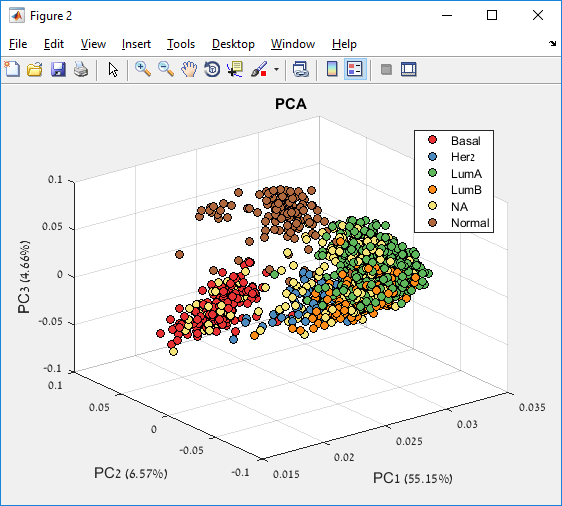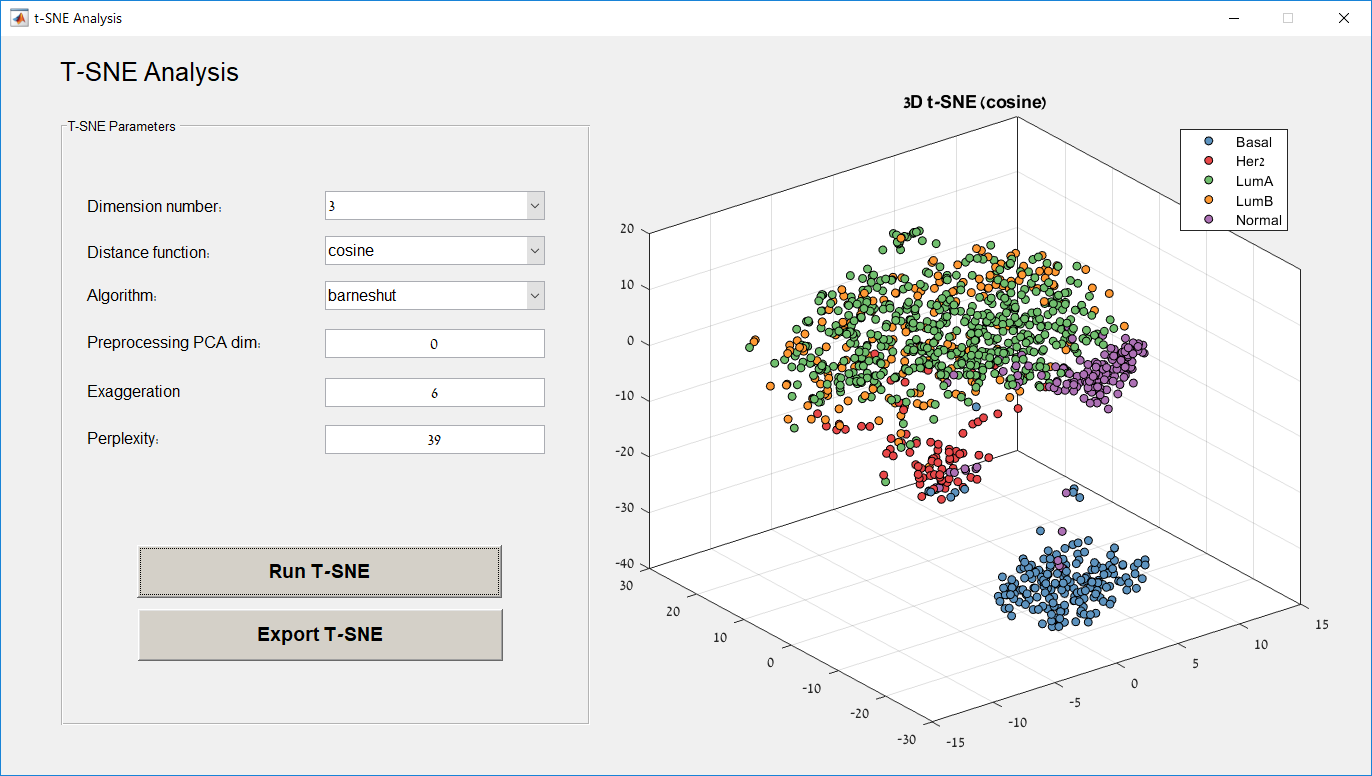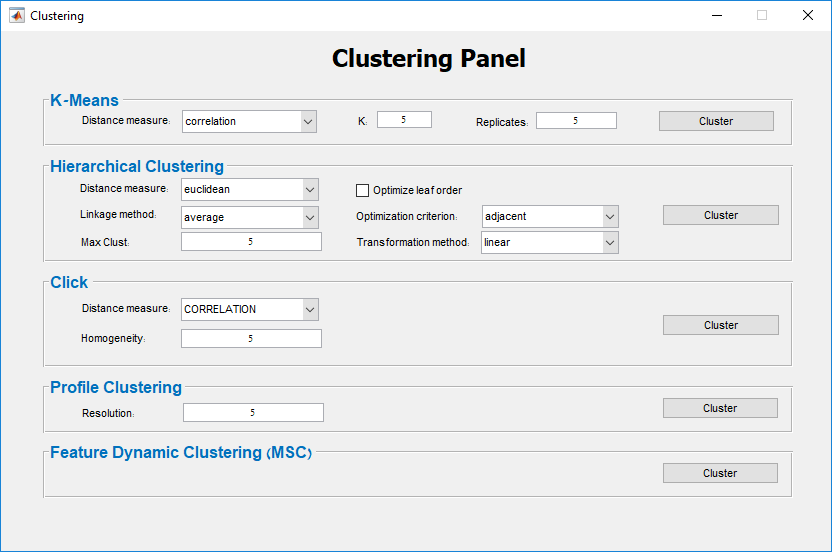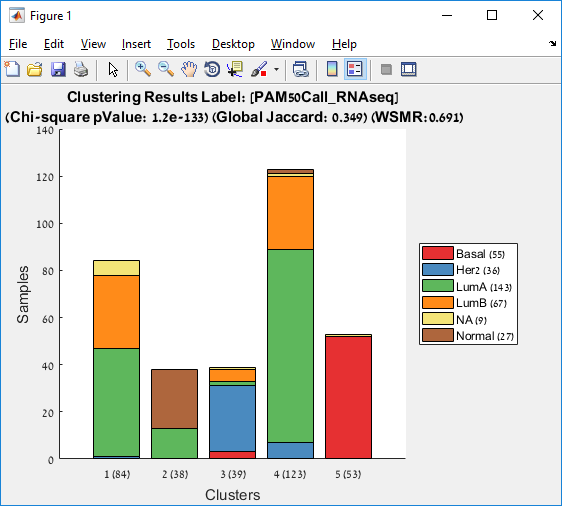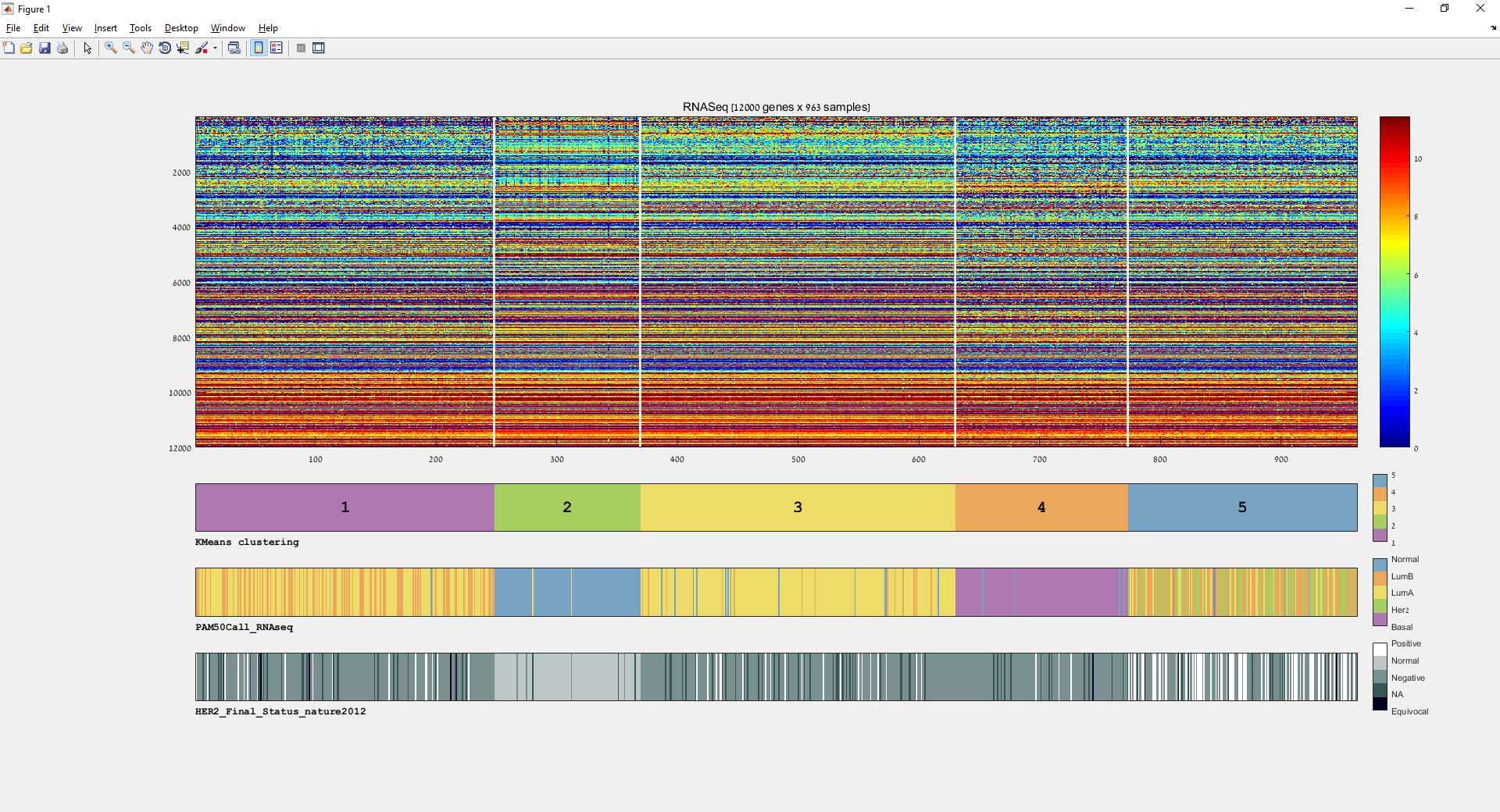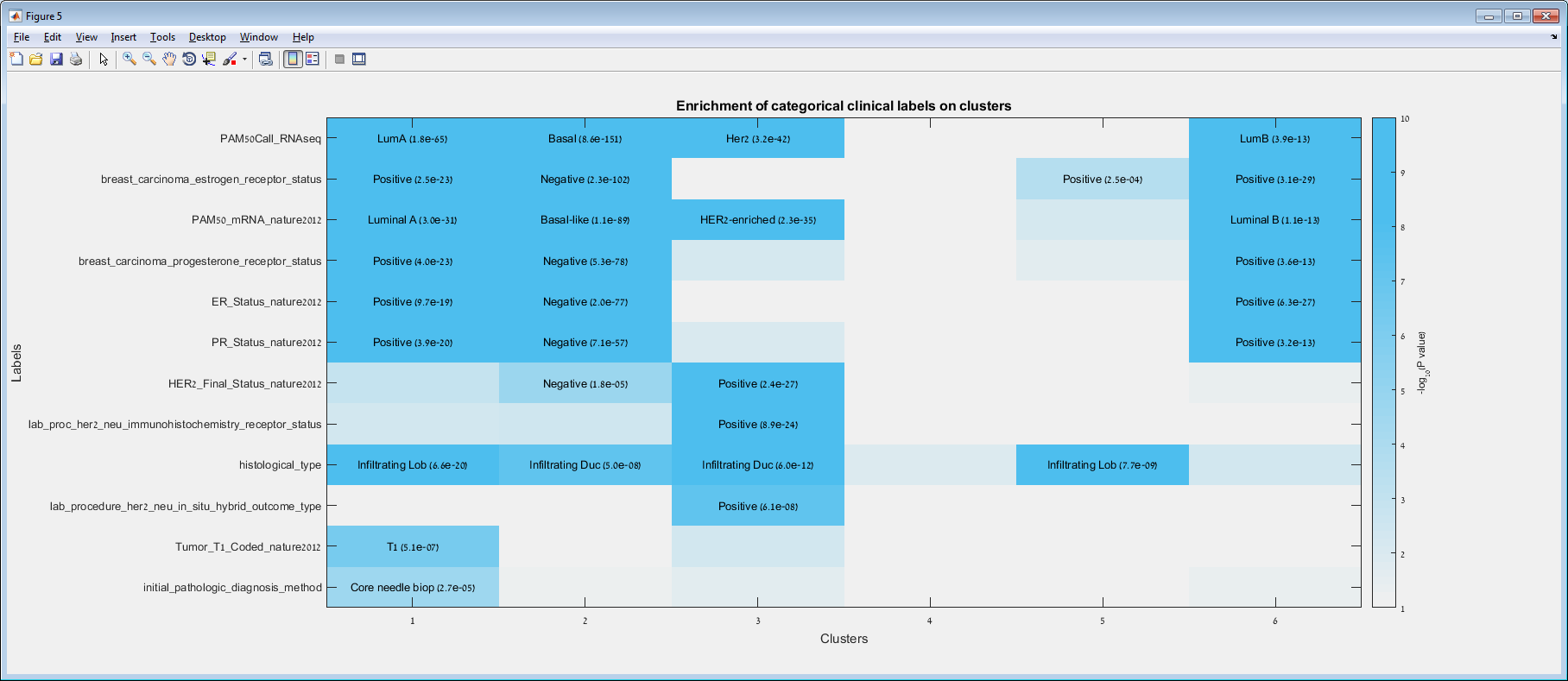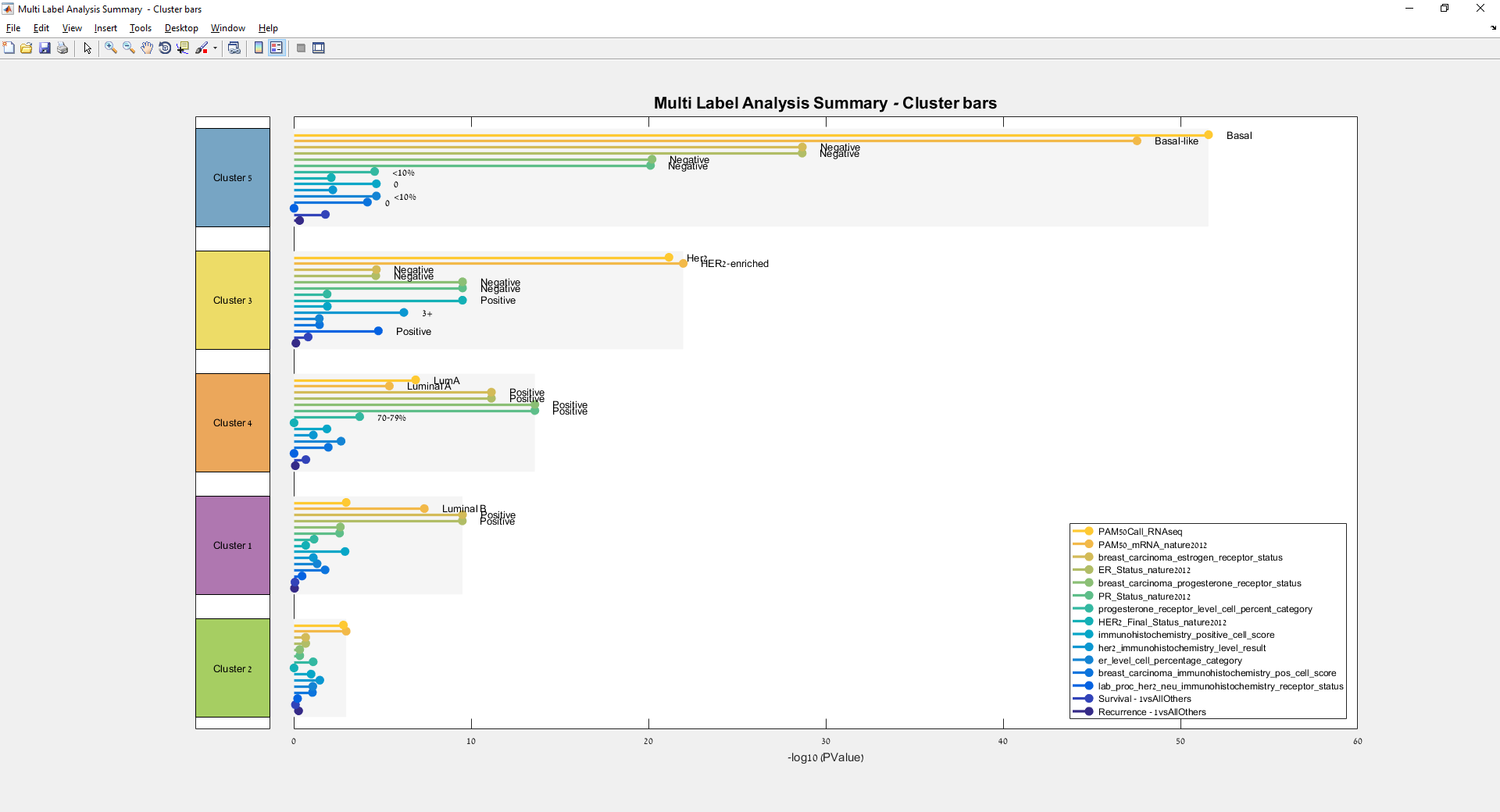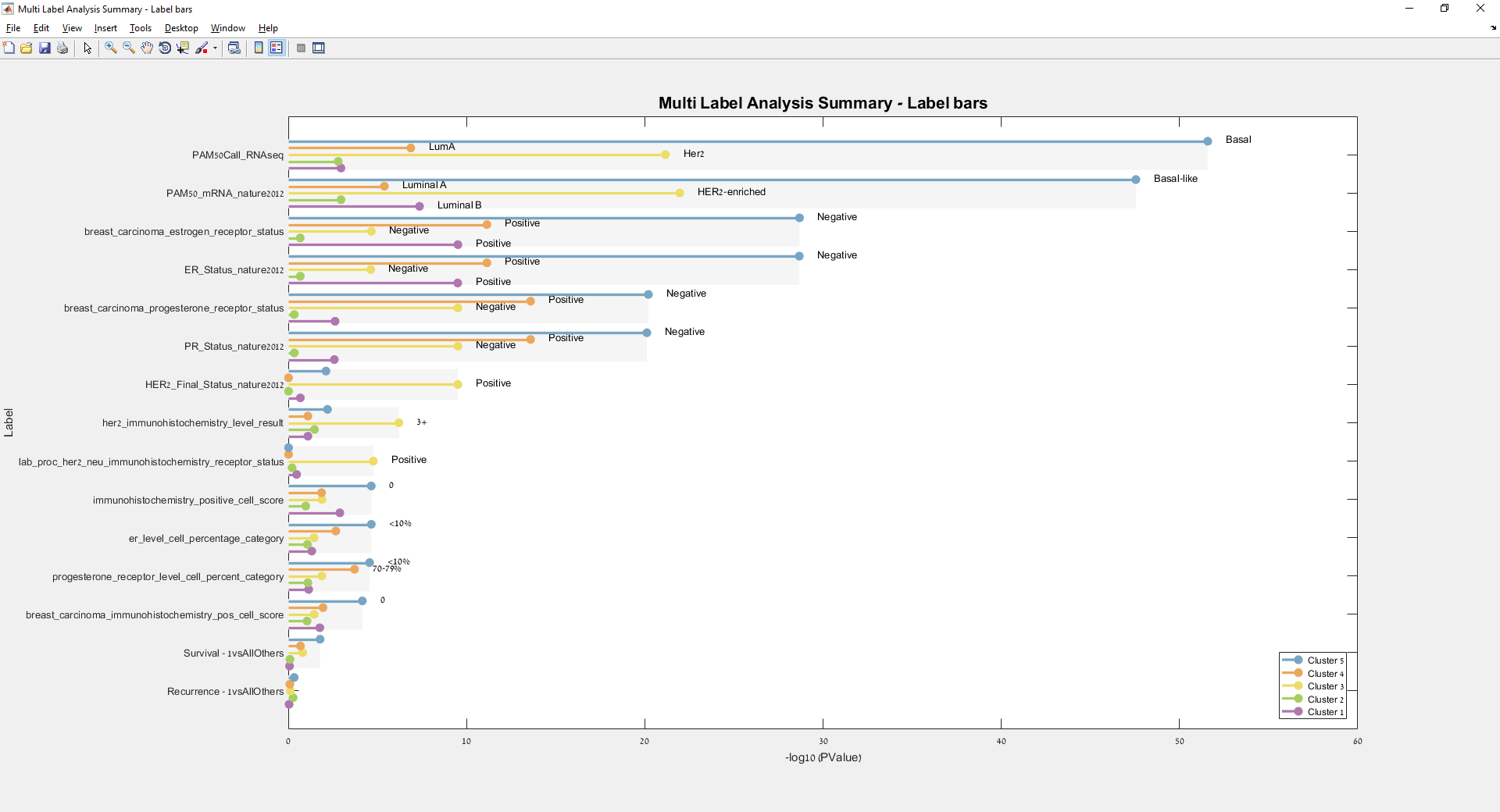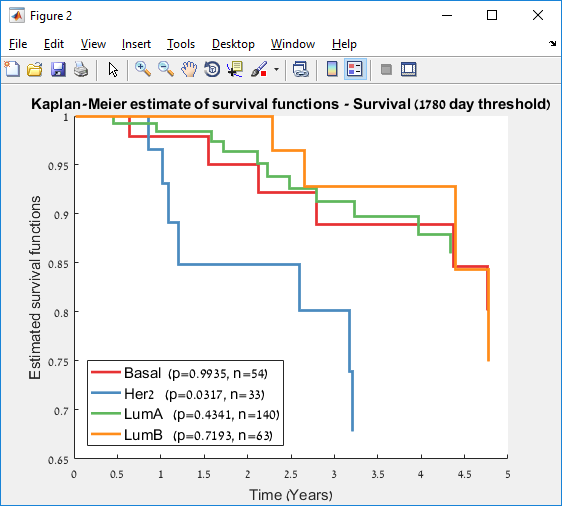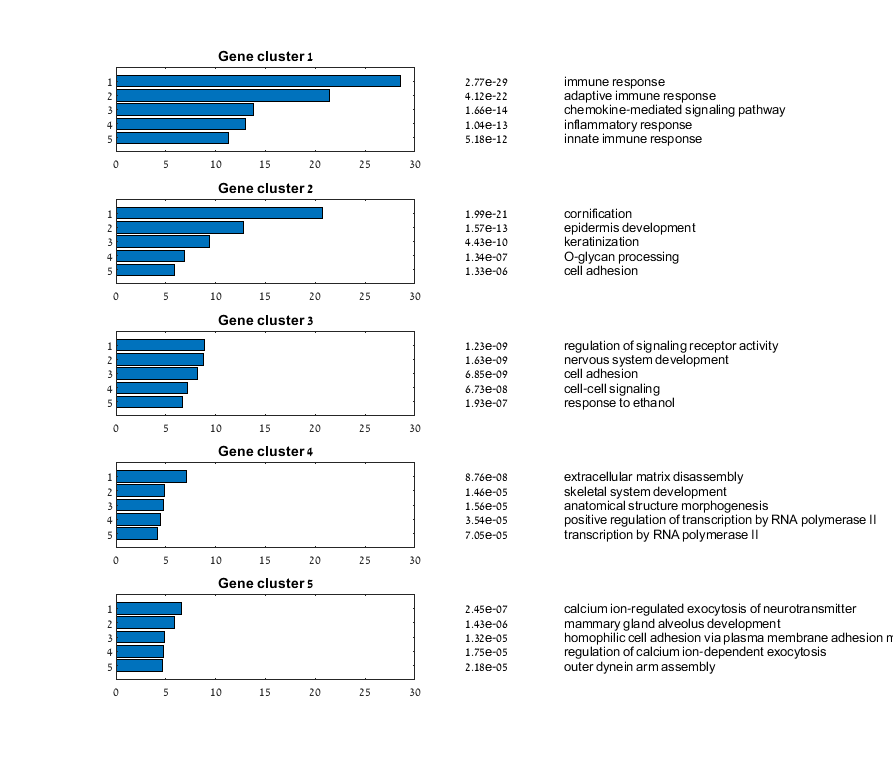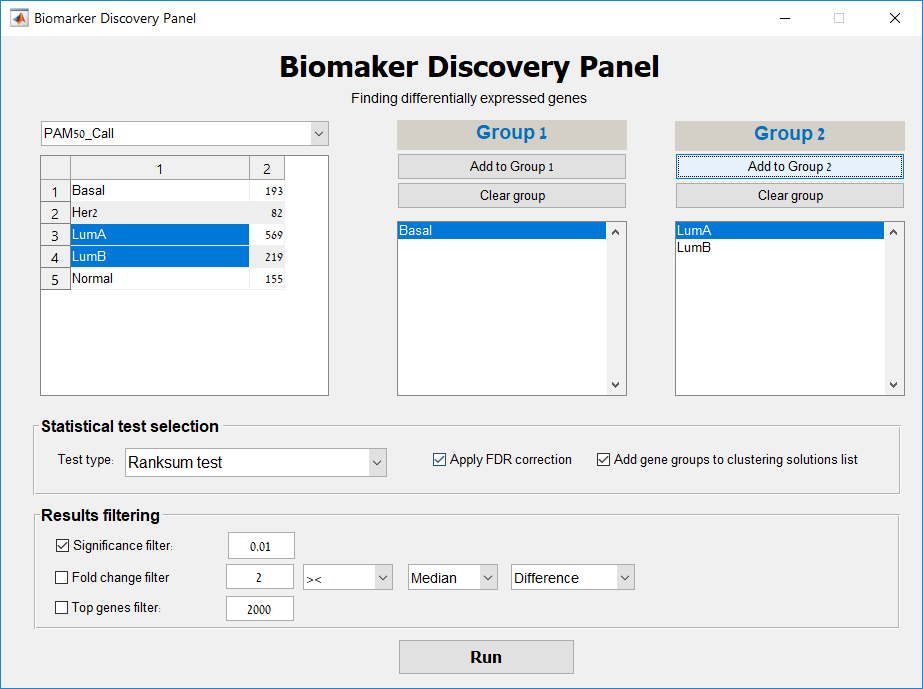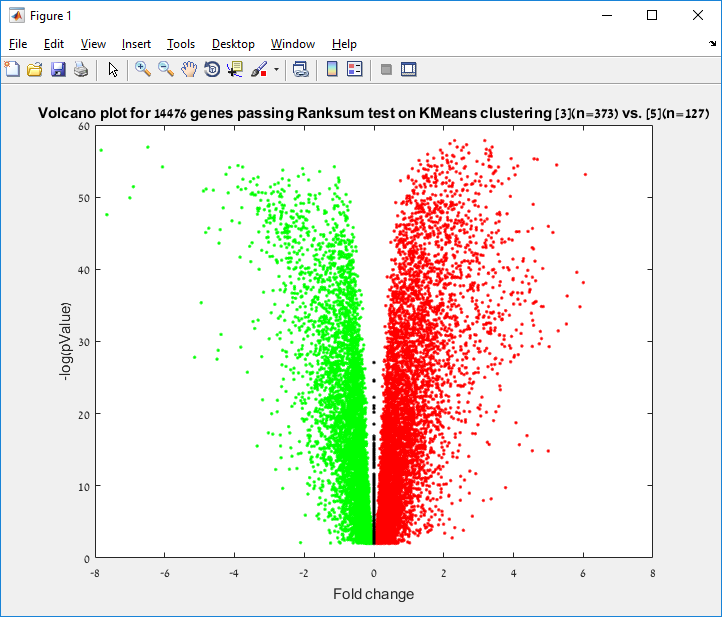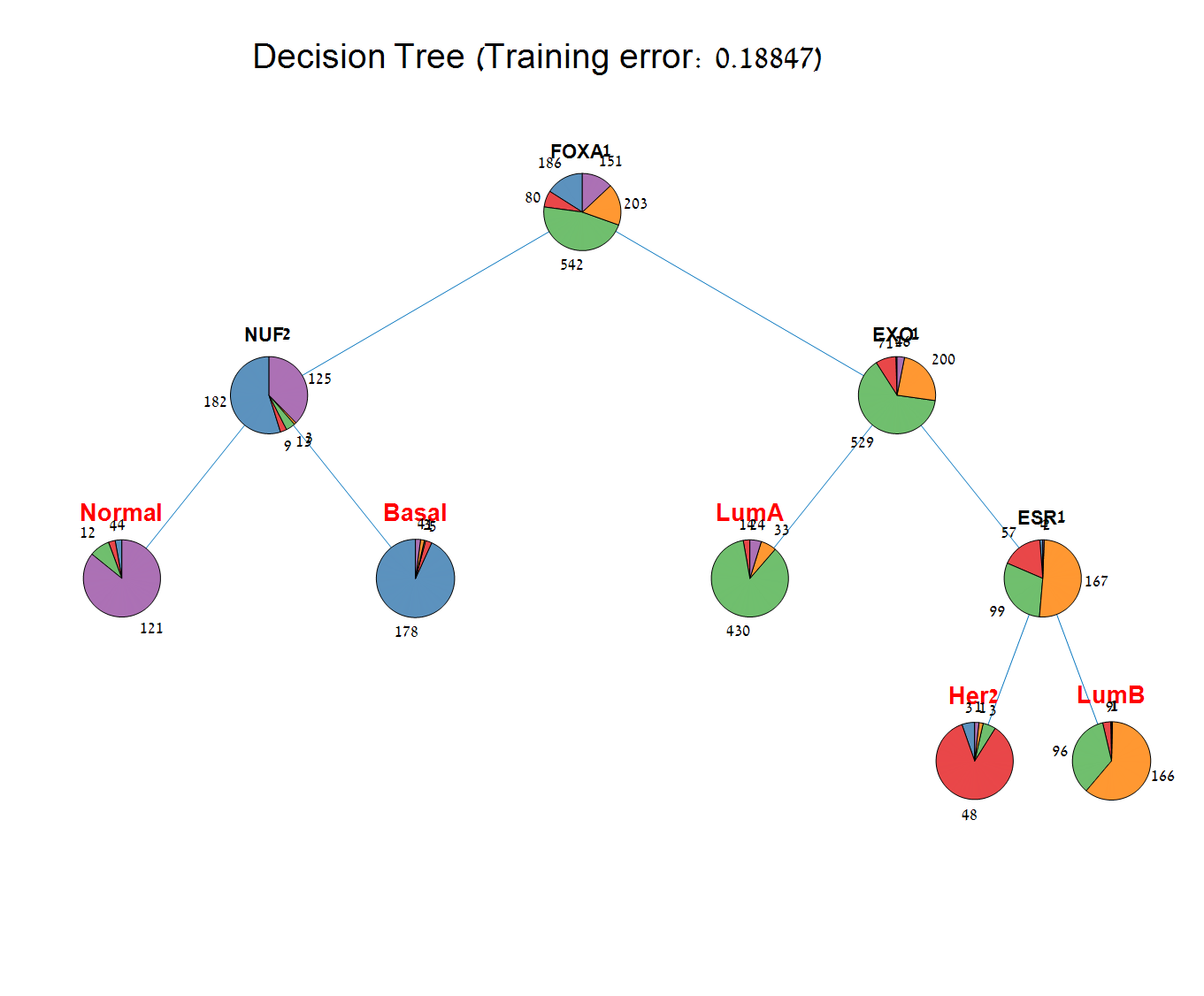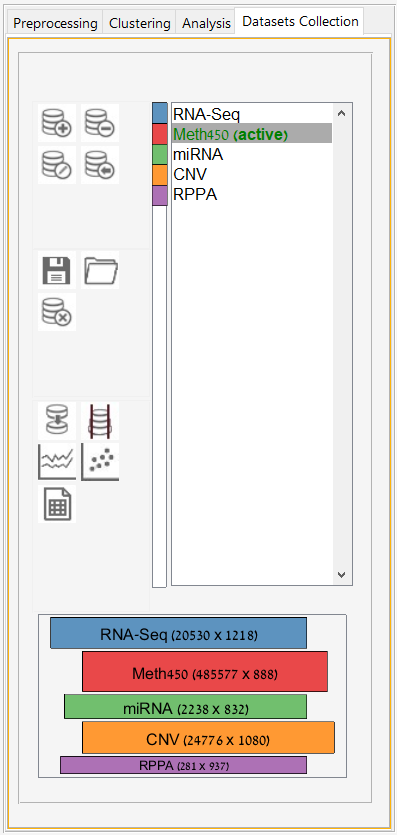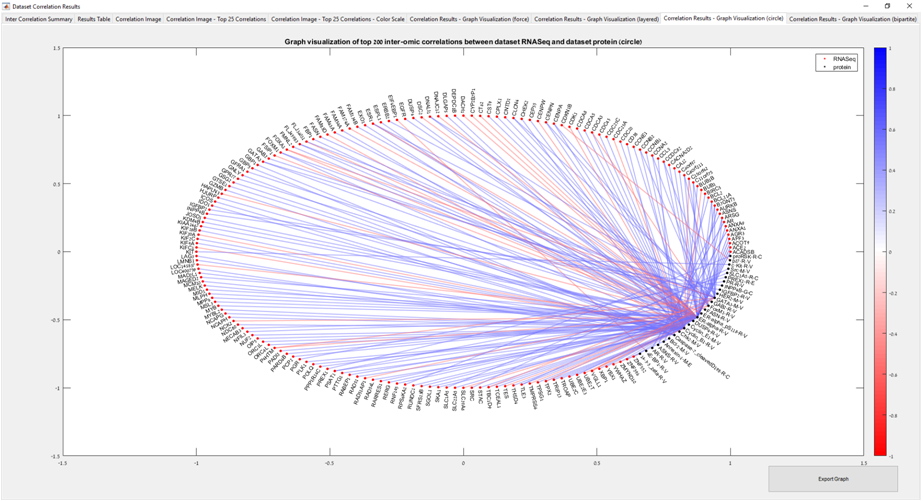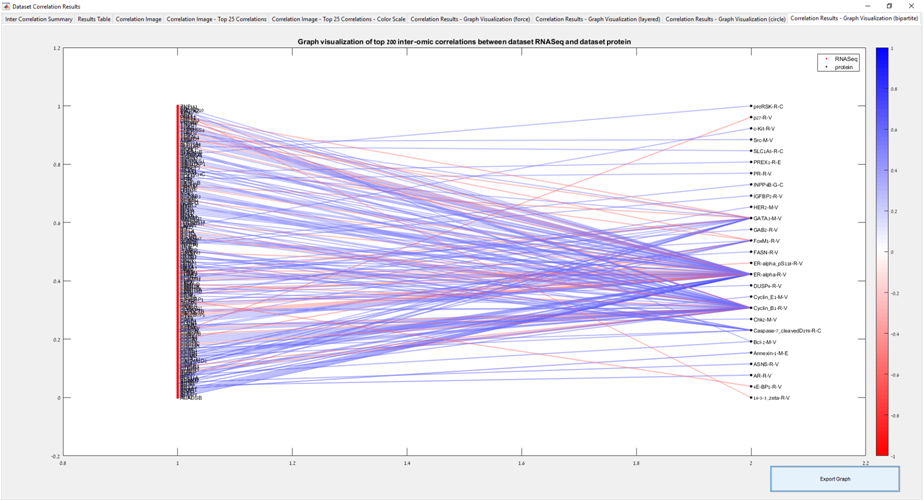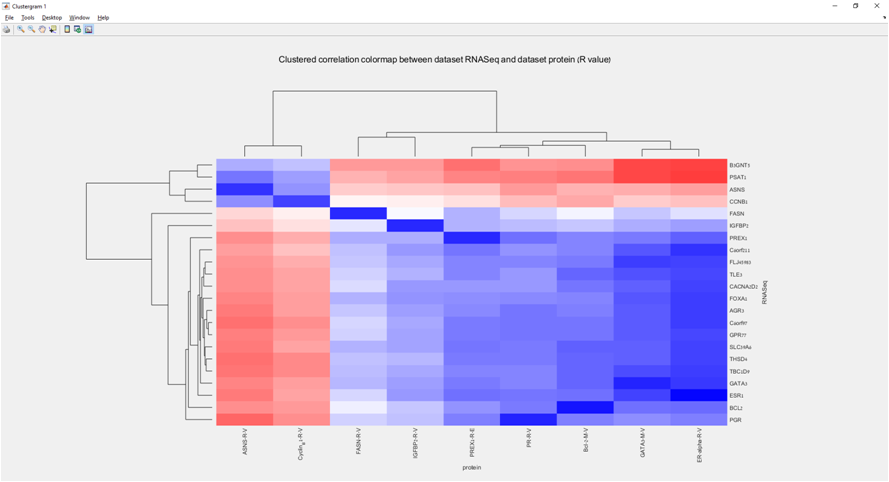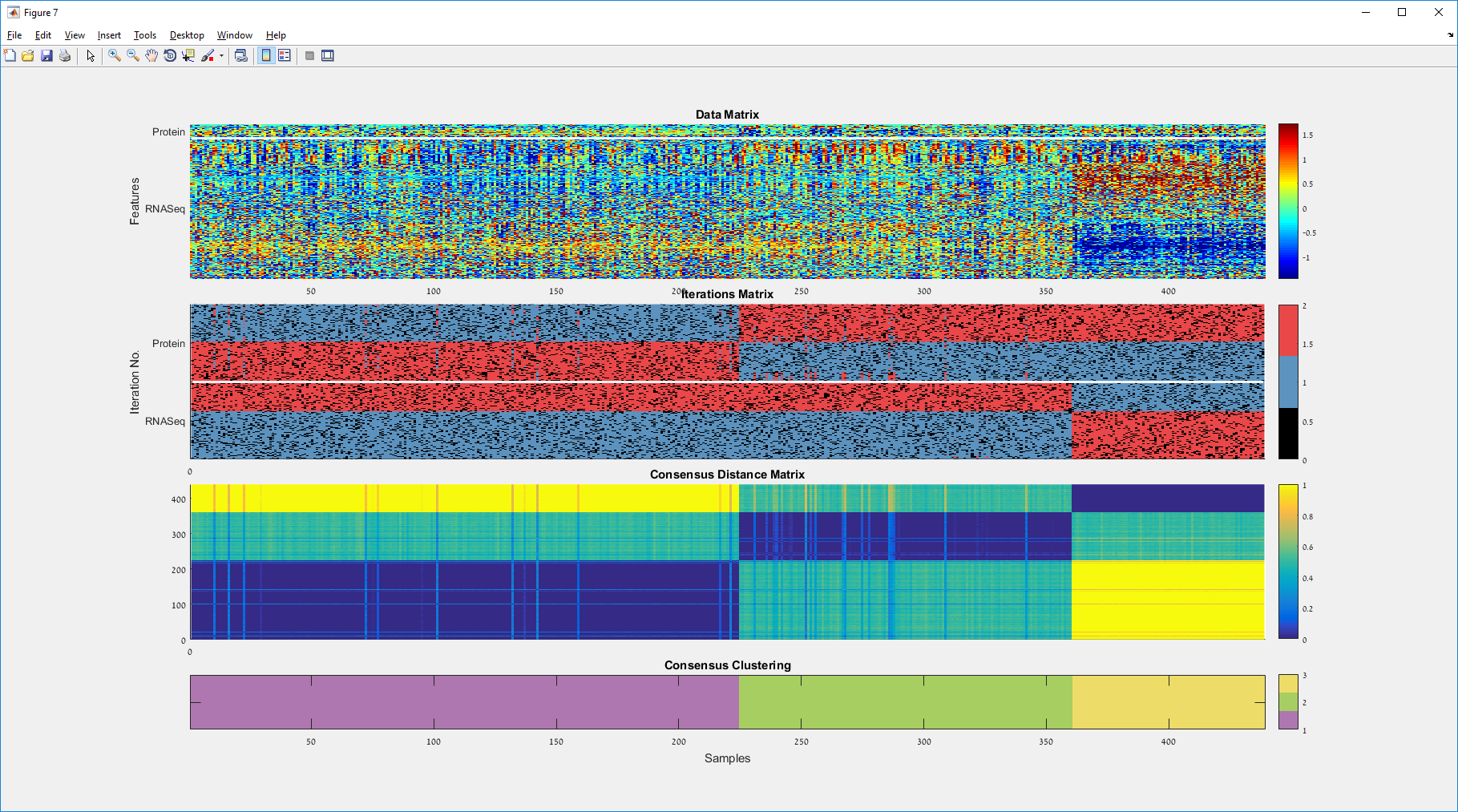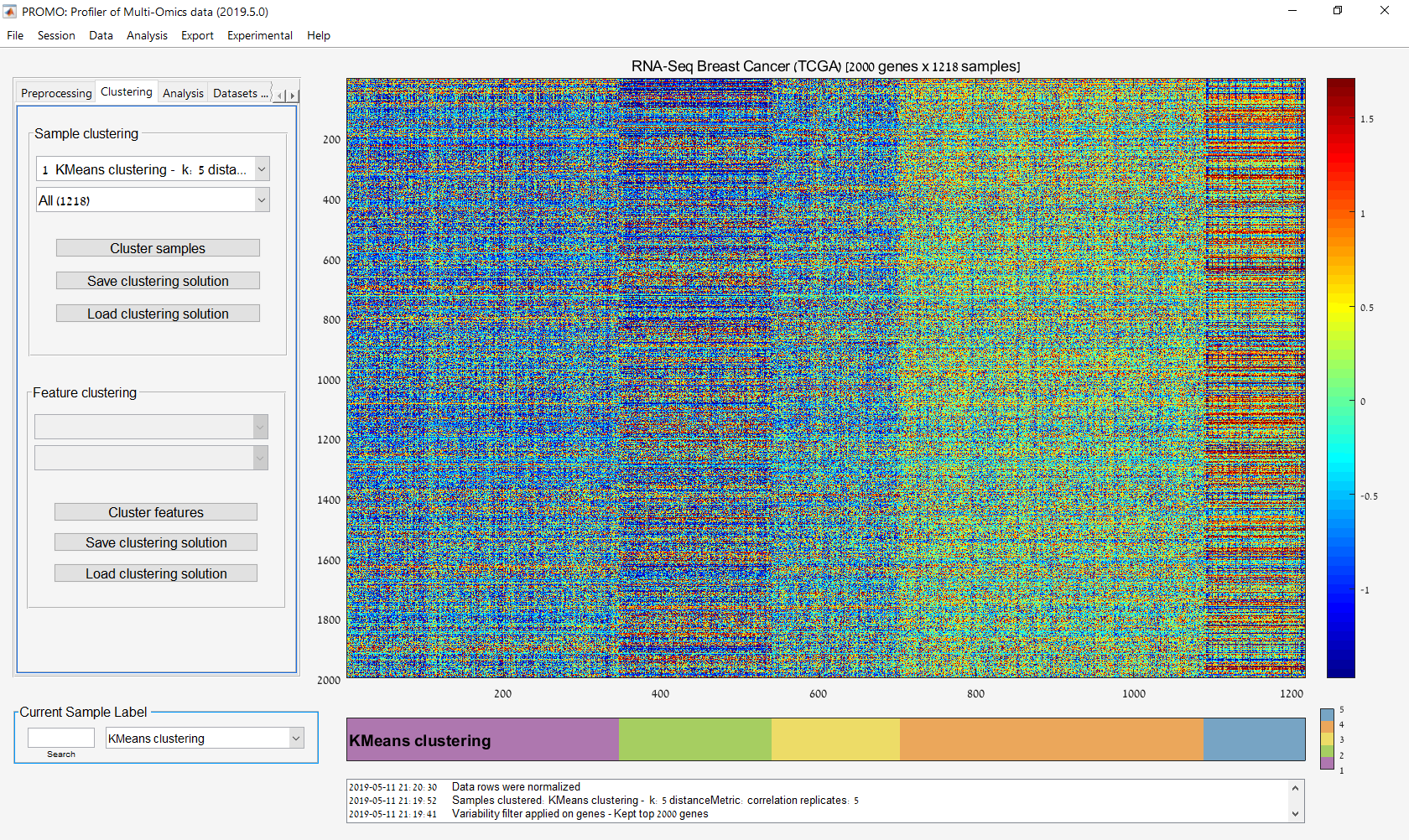
Main screen: An expression matrix whose samples (columns) are clustered using K-Means. The colorbar below the matrix shows the value of a selected label for each sample. Here the clustering label is shown. The log at the bottom of the screen documents the analysis work-flow and includes a description of every operation performed on the dataset.