| (1) | Human promoters. We constructed a set of putative promoters of known human genes by extracting sequences from the human genome that correspond to 1200 bp upstream of the genes' putative transcription start sites (TSSs) based on genes' start annotations (Human genome was downloaded from NCBI on July 2001). Human repetitive sequences are masked. The set contains putative promoters for 12981 human genes (we call it the '13K set'). The 13K set can be downloaded here (4MB, gzipped). |
| (2) | Models for BSs recognized by TFs. PRIMA uses the commonly used position weight matrices (PWMs) models for modeling binding sites recognized by TFs. In our analysis PWMs were obtained from the TRANSFAC database [3]. |
| (a) | Compute a similarity threshold T(P). Subsequences in the scanned promoters with similarity scores above this threshold are considered as 'hits' of P (i.e., putative binding sites of the TF modeled by the PWM). |
| (b) | Scan the promoters of the target and the background sets for identification of hits of P. |
| (c) | Employ a statistical test to examine whether hits of P are significantly over-represented in the target set with respect to the background set. |
Oct. '03: A new version of Prima which utilizes precomputed fingerprint files (for both Human and Mouse), and is, therefore, much faster, is now available as part of the EXPANDER package [4].
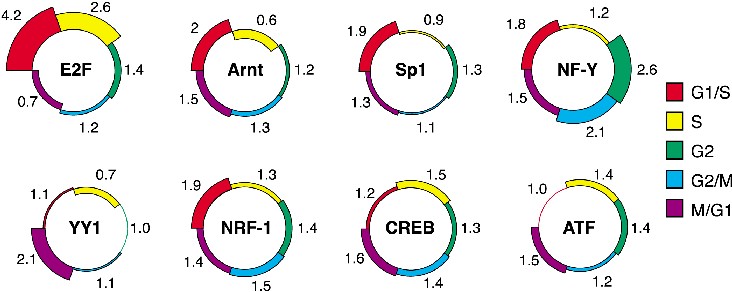
The fingerprint of a gene is the number of hits (putative binding-sites) of the various TFs that were identified in its promoter. The standalone version of PRIMA recomputes the fingerprints in each execution. While this allows more flexibility (e.g., in choosing the thresholds for declaring hits), this process is very time consuming. EXPANDER, on the other hand, executes PRIMA on a fixed set of precomputed fingerprints, which were constructed as follows: A set of about 17,000 human promoter sequences, spanning from 1000 bp upstream the TSS to 200 bp downstream the TSS, was scanned in order to locate putative BSs (hits). The scan was performed for each TF motif (PWM) in TRANSFAC (version 5.4, April '02) [3] that corresponds to a Human TF. The information on the number of hits of each PWM in a promoter is called the fingerprint of that promoter. The fingerprints of all human promoters are supplied with EXPANDER. The human promoter sequences were downloaded from Ensembl (release 13.30) [5]. Another set of fingerprints was prepared on mouse promoters (15,000 promoters, Ensembl release 13.30).
For most users we recommend using EXPANDER, both for promoter analysis and other computational and visualization tasks.
PRIMA is accessible via the "Made In Israel" bioinformatics portal.
| [1] | Elkon, R., Linhart, C., Sharan, R., Shamir, R., and Shiloh, Y.,
"Genome-wide In-silico Identification of Transcriptional Regulators Controlling
Cell Cycle in Human Cells",
Genome Research, Vol. 13(5), pp. 773-780, 2003. |
| [2] | Whitfield, M.L., G. Sherlock, A.J. Saldanha, J.I. Murray, C.A. Ball, K.E. Alexander, J.C. Matese, C.M. Perou, M.M. Hurt, P.O. Brown, and D. Botstein,
"Identification of genes periodically expressed in the human cell cycle
and their expression in tumors",
Mol Biol Cell, Vol. 13, pp. 1977-2000, 2002. |
| [3] | Wingender, E., X. Chen, R. Hehl, H. Karas, I. Liebich, V. Matys, T. Meinhardt, M. Pruss, I. Reuter, and F. Schacherer,
"TRANSFAC: an integrated system for gene expression regulation",
Nucleic Acids Res, Vol. 28, pp. 316-319, 2000. |
| [4] | EXPANDER - A Gene Expression Analysis and Visualization Software - http://acgt.cs.tau.ac.il/expander/expander.html. |
| [5] | The Ensembl Project - http://www.ensembl.org. |