EXPANDER has a "ChIP-Seq Analysis" menu item which provides the following tools for the joint analysis after loading both ChIP-seq and gene expression data:
1) Examining
gene expression distribution within ChIP-Seq-associated genes: this is done via
ChIP-Seq Analysis >> Integration with expression data >> ChIP-Seq
vs. GE analysis. Upon selecting this option, the following dialog box will
appear:

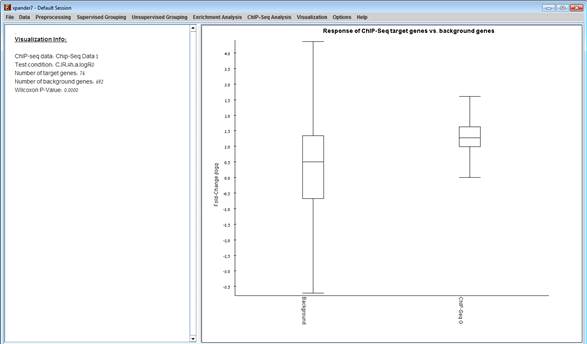
In the dialog box select the relevant ChIP-seq data set name, the base-condition and the test-condition, and click OK. A box plot showing the distribution of test-base GE ratios will be displayed for the set of chip-seq genes (right) and the rest of the genes in the gene-expression data set (left).
2) Extracting a grouping solution from ChIP-seq-gene-groups intersection: this is done via ChIP-Seq Analysis >> Integration with expression data >> ChIP-Seq Intersection. Upon selecting this option the following dialog box is displayed:

In the dialog box select the relevant grouping solution and ChIP-seq data set name, and click OK. A grouping solution visualization tab will be added to the main window. It will contain a gene-group for each non-empty intersection between a gene-group in the original solution and the chIP-Seq genes. Visualization is similar to clustering visualization.
3) Gene Set Enrichment Analysis: this is done via ChIP-Seq Analysis >> Integration with expression data >> GSEA. ChIP-Seq data can be selected as "Grouping solution". For further information please refer to: Gene Set Enrichment Analysis (GSEA) .
4) ChIP-Seq Enrichment: this is done via ChIP-Seq Analysis >> Integration with expression data >> ChIP-Seq Enrichment. For further information please refer to: ChIP-Seq Enrichment Analysis .