Expander operates on two types of data:
a) Expression data – can be either cDNA microarray data (expected as log 2 (R/G)) values OR Oligonucleotide array data (expected as positive expression levels).
If one wishes to perform functional analysis or promoter analysis, an ID conversion file should be loaded along with the data file. The conversion file maps each probe ID (first column) in the data file into a corresponding conventional gene ID that is used in the GO annotation and TF fingerprint files that are supplied with EXPANDER.
b) Gene sets data – contains predefined sets of genes. In this data
type, the conventional gene IDs that are used by EXPANDER in the GO annotation
and TF fingerprint files are expected.
For details regarding the Gene ID convention that is used for each organism, refer to the Supplied files section.
For details regarding the data files formats see the File Formats section.
Loading gene expression data:
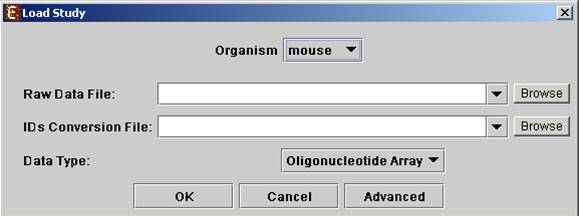
Select the New Session option from the File menu. From the submenu select Expression Data.
The following dialog box will appear:

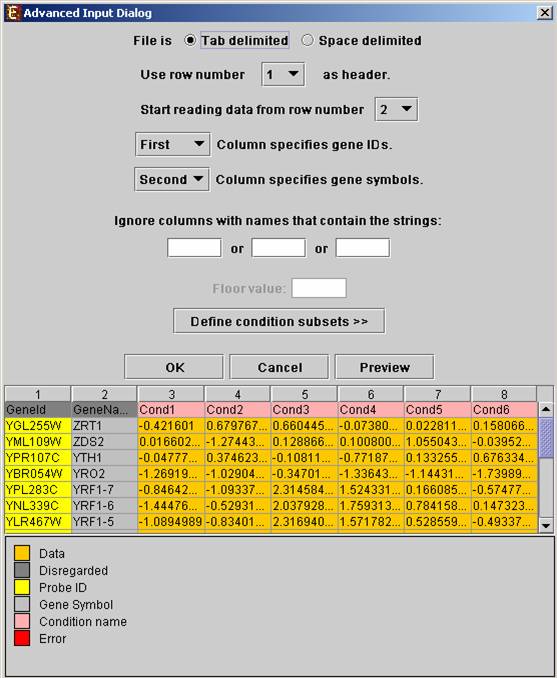
Upon pressing the “Advanced” button after filling the “Raw Data File” field, an “Advanced dialog box” appears. This dialog box can be used in order to facilitate the data load of files that are not in the required format. It also enables the user to input a floor value, to which all entries that are below that value will be set (this option is available only for oligonucleotides data). The first few rows and columns of the data are displayed in a table, demonstrating the way the data is read by the program according to the current input values.
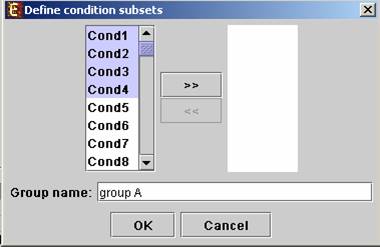
Upon pressing the “Define condition subsets” button, a dialog box appears, enabling the grouping of conditions under a common subset name. This partition is used for visualization purposes.
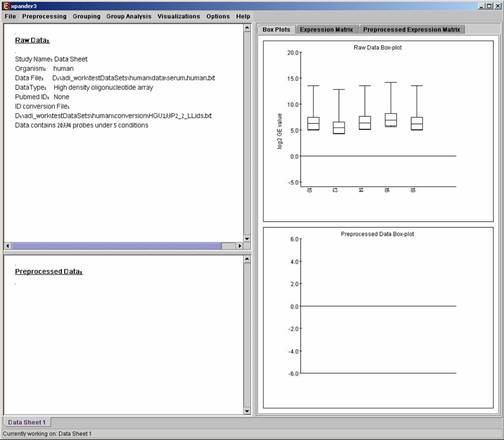
After loading a gene expression data set, a “Session Data” display tab is added to the main window (see example below). It contains information regarding the raw data file, a box plot chart, and an expression matrix visualization of the raw data.
Loading gene sets data:
Select the New Session option from the File menu. From the submenu select Gene Sets.
The following dialog box will appear:
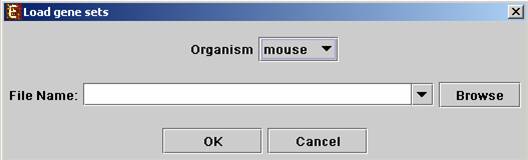
For details regarding the data files formats see the File Formats section.
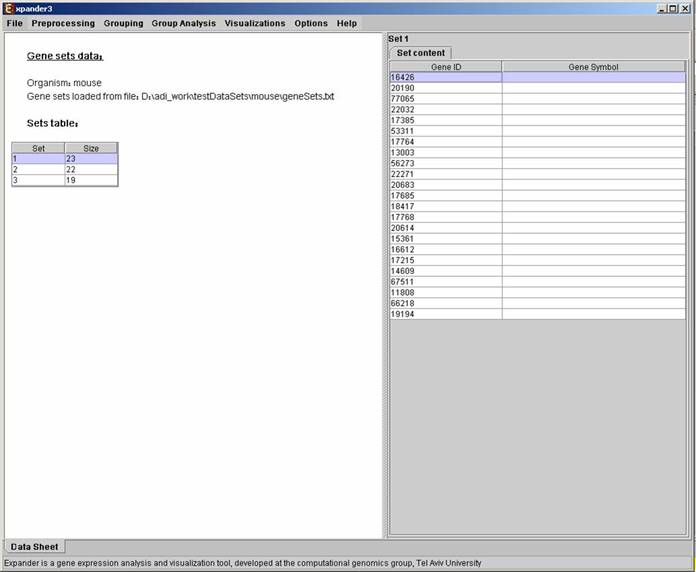
After loading a gene sets data, a “Session Data” display tab is added to the main window (see example below). It contains information regarding the data file, and a table describing the different sets (set number and size). Upon clicking on a row in the table, the corresponding gene list appears on the right.